
Finding the recipe for life on Earth
Three key factors were required for life to develop on Earth – but which factor came first? Recent research could help settle the…

We all know that DNA → RNA → protein. But did you know that some genes don't encode proteins but rather RNAs with important cellular functions?
According to the central dogma of molecular biology DNA is used as a template to create messenger RNA (mRNA), which in turn is translated into proteins that build the tissues in our bodies and carry out the main functions of our cells and organs. In other words, DNA → mRNA → proteins. Interestingly, though, only around 2% of the DNA in our whole genome codes for proteins! So, what does the other 98% of the human genome do? Much of it may not actually have a specific function. However, in the latter part of the 20th century and the early 21st century, some of this non-coding DNA has been shown to not only contain important regulatory elements for transcription, but also sequences that encode various non-coding RNAs (ncRNAs) that have functions in many cellular mechanisms.[1]
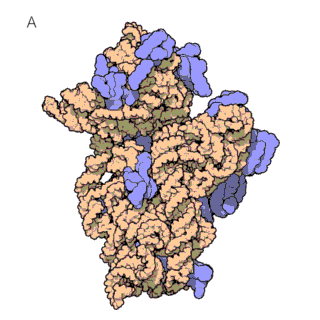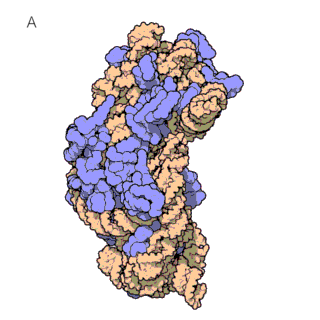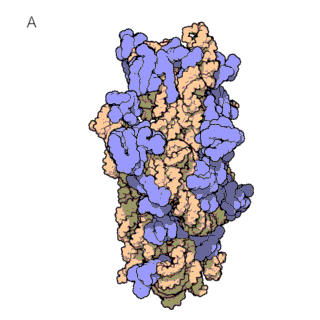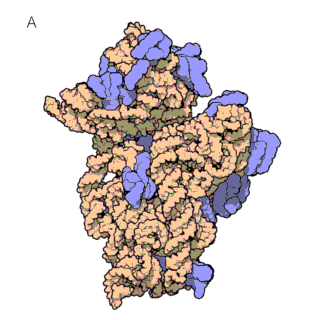
The first known classes of non-coding RNAs were ribosomal (rRNAs) and transfer RNAs (tRNAs), discovered in the 1950s. tRNAs are essential for protein synthesis, as they decode the mRNA sequence and translate it into an amino acid sequence. This translation of mRNAs into proteins occurs on the ribosome, which itself consists of large ribosomal RNAs and multiple ribosomal proteins (figure 1a and 1b). In fact, ribosomes are so important for the cells that rRNAs are the most abundant RNAs in the cell!
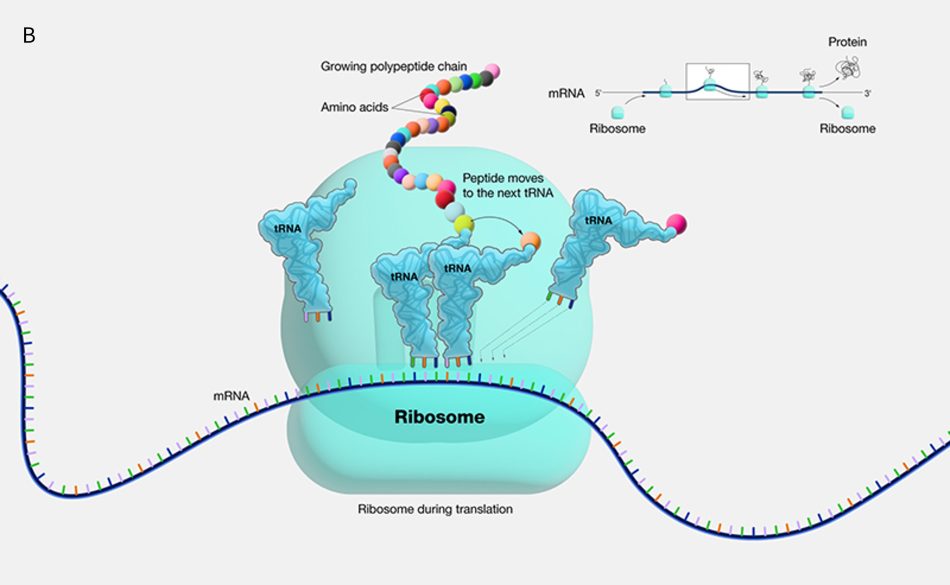
Both tRNAs and rRNAs have complex 3D folded structures that are essential to their function, much like proteins.
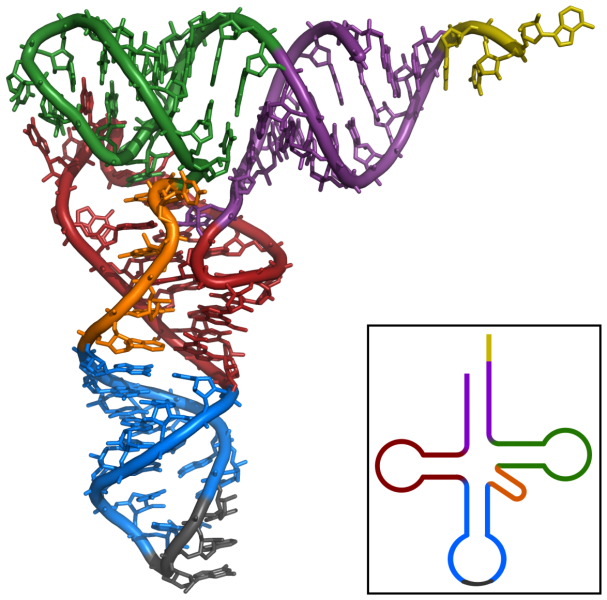
rRNAs and tRNAs form a group of so-called housekeeping ncRNAs because they are necessary to maintain fundamental cellular processes. A second group of non-coding RNAs was only discovered later in the 1990s. These RNAs, called regulatory ncRNAs, have diverse regulatory roles in the cell. Non-coding RNAs of less than 200 base pairs are called small non-coding RNAs (sncRNAs), while all non-coding RNAs longer than 200 base pairs are long non-coding RNAs (lncRNAs; figure 3), and both can be further divided into groups based on their function.[2]
Although they only make up a small fraction of the genome, we understand now that non-coding RNAs are important for various cellular processes. But how do these RNAs fulfil their functions? In most cases, ncRNAs must interact with other molecules. For example, rRNAs interact with proteins during protein synthesis; however, it seems they can also work with almost any kind of molecule. RNAs can even interact with other RNAs. One of the most exciting examples of RNAs interacting with each other are microRNAs, which are found in almost all organisms apart from bacteria and some yeast species. MicroRNAs consist of only 21–23 nucleotides and are described as having ’hairpin’ structures because, before processing, they fold back on themselves like a hairpin. Despite their tiny size, they act as a guide for powerful scissors inside the cell by binding to a protein called RNA-induced silencing complex (RISC), which cuts up coding mRNAs. When the microRNA-bound RISC meets an mRNA with a complementary nucleotide sequence, it cuts the mRNA and prevents its translation into a protein (figure 4). Nowadays, scientists routinely use this RNA-silencing mechanism in their experiments to study how cells behave if a particular protein is removed.[3]
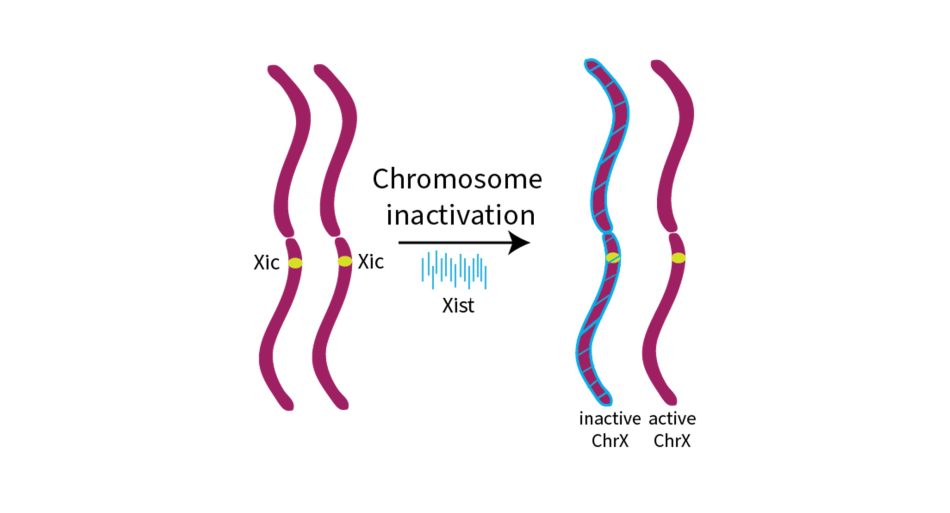
Another fascinating role of RNA has been discovered in the chromosomes of mammals. Human cells contain 23 pairs of chromosomes. For each chromosome pair, we receive one chromosome from our mother and one from our father. This means that we have two copies of each gene and normal cell function is based on the amount of protein that will be translated from the two gene copies. If a cell ends up with more or fewer than two of a particular chromosome, it can lead to serious health problems, such as Down Syndrome, which is caused by three copies of chromosome 21. However, there is one special pair of chromosomes that doesn’t always match: the sex chromosomes, which may be present as two X chromosomes in female cells or one X and one Y in male cells. But how does the cell handle the different numbers of X chromosomes in different individuals? This problem is solved by setting the correct number of expressed X chromosomes to one in both sexes. In early development, the non-coding RNA Xist coats one of the X chromosomes in XX individuals and inactivates it (figure 5). In this way, female XX cells have as many active X chromosome genes as male XY cells.[4]
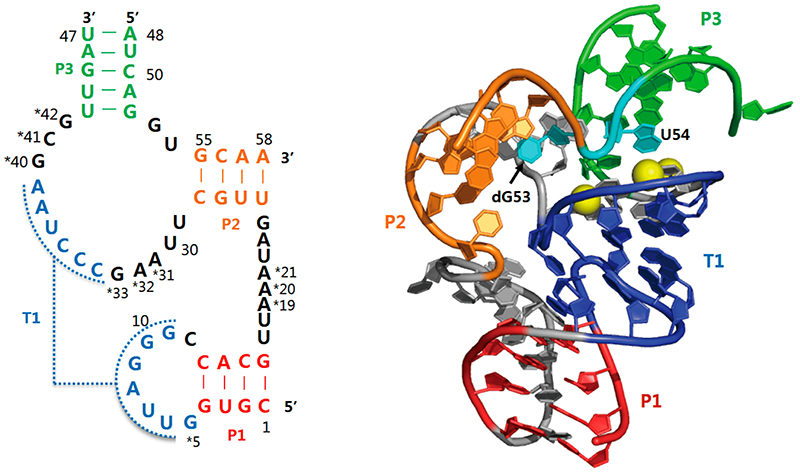
These examples of RNA’s functions show the incredible complexity of our cells. How did this complexity evolve? In 1986, Walter Gilbert proposed that the first biomolecular processes were completely based on RNA, long before DNA and proteins evolved. This concept is called the ‘RNA World’ hypothesis.[5] There are a number of arguments supporting this hypothesis. We already know that RNA molecules can carry genetic information just like DNA, although RNA degrades more easily so DNA provides more stable storage. However, some organisms, such as certain viruses, use the instability of RNA to their benefit by storing their genomes as RNA, which is more liable to mutate and thereby enables fast adaptation. Moreover, there are special RNA molecules called ribozymes, which have a folded structure like microRNAs but additionally possess a catalytic function like proteins: they can cut proteins and RNA molecules or accelerate enzymatic reactions. This means that although the fundamental principle of molecular biology is DNA à RNA à protein, RNA can fulfil the functions of all three units! The most famous ribozymes found in nature are the hammerhead ribozyme and hairpin ribozyme (figure 6). They were first identified in plant viruses,[6] where they are used to cut RNAs and even produce tiny infectious particles.[7] However, researchers have also found them in human cells, where they’re probably involved in mRNA processing.[8] In the future, we might even be able to use them to treat cancer and other diseases.[9]
Another function that biomolecules based on RNA can perform in cells is mediating signalling within or between cells. Proteins can perform this role but there are also a number of small signalling molecules, and some very important ones, such as cyclic AMP and cyclic di-GMP, are made from RNA building blocks.[11] Together with ribozymes, these molecules might represent the last remnants of the RNA World!
Sadly, while these remnants in our cells are fascinating to think about, it’s incredibly difficult to collect direct evidence for or against the RNA World, which would have existed around 4 billion years ago.
We are only starting to reveal the functions and importance of RNA molecules in cellular processes. My own research project explores the function of RNAs on the surface of chromosomes during cell division. Other research groups are exploring the role of ncRNAs in diseases and how to use them in medical treatments.[12] Scientists have only recently finished sequencing the 3.2 billion nucleotides of the human genome[13] and there is still a great deal of work to do to understand it.
The story of non-coding RNAs shows how we thought we understood the genome, but then realized that nature is still much more complex than we ever expected. It is crucial to stay curious and have an open mind to notice all the unexpected and beautiful things hidden behind nature’s complexity.
Editor’s note: Some parts of the introduction and conclusion were rephrased to avoid any misunderstanding concerning the nature of ‘junk DNA’, which is not the focus of this article. The title was also modified to better reflect this.
DNA: Deoxyribonucleic acid sequence carries the genetic information of the organism inside each cell
RNA: Ribonucleic acid is a less-stable nucleic acid than DNA, but it fulfills various important functions in the cell
mRNAs: Messenger RNAs serve as a transcript of the DNA and a template for its translation into proteins
tRNAs: Transfer RNAs decode the mRNA template into an amino acid sequence to make proteins into ribosomes
rRNAs: Ribosomal RNAs are the most abundant RNA species in the cell; together with ribosomal proteins they make up the structural components of the ribosomes
ncRNAs: Non-coding RNAs are RNA molecules that are not translated into proteins. These include tRNAs and rRNAs, as well as regulatory ncRNAs
Ribozymes: These RNAs have a catalytic function like enzymes; not to be confused with ribosomes.
[1] A BBC Future article on the role of the ‘dark genome’: https://www.bbc.com/future/article/20230412-the-mystery-of-the-human-genomes-dark-matter
[2] Palazzo AF, Lee EW (2015) Non-coding RNA: what is functional and what is junk? Frontiers in Genetics 6: 2. doi: 10.3389/fgene.2015.00002
[3] Filipowicz W et al. (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Current Opinion in Structural Biology 15: 331–341. doi: 10.1016/j.sbi.2005.05.006
[4] An article on the inactivation of the X chromosome on the UCLA News website: https://newsroom.ucla.edu/releases/rna-silences-x-chromosome
[5] Higgs P, Lehman N (2015) The RNA World: molecular cooperation at the origins of life. Nat Rev Genet 16: 7–17. doi: org/10.1038/nrg3841
[6] Fedor MJ (2000) Structure and function of the hairpin ribozyme. J Mol Biol 297: 269–291. doi: 10.1006/jmbi.2000.3560
[7] Hammann C et al. (2012) The ubiquitous hammerhead ribozyme. RNA 18: 871–885. doi: 10.1261/rna.031401.111
[8] de la Peña M, García-Robles I (2010). Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep. 11: 711–716. doi: 10.1038/embor.2010.100
[9] An article on the trans-splicing ribozyme as genetic therapy on Advanced Science News: https://www.advancedsciencenews.com/therapeutic-applications-group-intron-based-trans-splicing-ribozymes/
[10] Lee K-Y, Lee B-J (2017) Structural and Biochemical Properties of Novel Self-Cleaving Ribozymes. Molecules 22: 678. doi: 10.3390/molecules22040678
[11] Nelson JW, Breaker RR (2017) The lost language of the RNA World. Sci Signal 10: eaam8812 doi:10.1126/scisignal.aam8812
[12] Bhatti GK et al. (2021) Emerging role of non‐coding RNA in health and disease. Metab Brain Dis 36: 1119–1134. doi: 10.1007/s11011-021-00739-y
[13] A BBC Future article on the first sequencing of a complete human genome: https://www.bbc.com/future/article/20230210-the-man-whose-genome-you-can-read-end-to-end
This article relates to the curriculum topics of Molecular biology, Cell biology, and Nucleic acids and could be used for stretching more capable students beyond the curriculum.
Comprehension questions:
A nice extension could be to run a classroom debate about which molecule is the most important in the cell – DNA, RNA or protein?
Kathy Freeston, Biology teacher, East Leake Academy, UK

Three key factors were required for life to develop on Earth – but which factor came first? Recent research could help settle the…
Stroll through biological databases: Walking on chromosomes is a CusMiBio project that teaches students how to explore biological databases and…

Play the part: students take on the roles of different components of a synapse to act out synaptic transmission and learn about neurobiology.