Evolution in action: the 67 000-generation experiment Understand article
A unique experiment tracks microbes changing over thousands of generations – so we can watch evolution on fast-forward.
As an evolutionary biologist, I like to think that studying evolution is akin to studying the stars. No astronomer lives long enough to observe a star being born, getting old and dying. The Sun, for example, has a life cycle of about 10 billion (1010) years – a timescale out of all proportion to a human life. But because the Universe is full of stars, with millions of them in the Milky Way alone, astronomers can observe many stars at different stages of their life cycle. From these observations, they can gather enough data to work out the different stages of a star’s life and map their whole evolution.
Evolutionary biologists live no longer than astronomers, so no-one has seen a single vertebrate species diverging and evolving into two or more different species. But like astronomers, we can exploit the fact that there are millions of species on Earth that split from others at different points in time, and we can access the records of this history preserved in rocks and in the genetic material of organisms, either dead or alive.

Paul Williams / Flickr
In this context, evolutionary geneticists who study micro-organisms have an advantage over biologists concerned with the evolution of more complex species such as vertebrates: the microbes reproduce and mutate so rapidly that they put evolution on fast-forward. In this article, we look in detail at an extraordinary experiment that has tracked the evolution of the bacterium Escherichia coli over an amazing 67 000 generations. Translated into human generations, this number is equivalent to around one million years – which would take us back to well before the beginning of our own species, Homo sapiens. Using E. coli, which replicates itself six or seven times each day, it has all happened in less than 30 years.
But how can tracking populations of micro-organisms tell us anything about the evolution of other species, or even of humans themselves? First, let’s have a look at what we mean by evolution.
What is evolution?
Evolution is any change in the frequency of genetic variants (alleles) within a group of organisms of the same species (population) from one generation to the next. There are several mechanisms that can cause evolution, including migration (when a group of new individuals join or leave an existing population) and simple chance (when individuals have unequal opportunities to reproduce). The best-known mechanism, which is often mistaken for evolution itself, is positive natural selection: the mechanism by which individuals with some combinations of alleles in a species within a given environment produce more offspring than individuals with other combinations, leading to adaptations in that species.

Marcin Bajer / Flickr
Many changes in allele frequency do not lead to the emergence of a new species – but when a split occurs, this is always the consequence of many different genetic variations accumulating in separate populations. Studying such fundamental mechanisms of evolution – how genetic changes arise and then spread through or disappear from a population, and how they contribute to an organism’s ‘fitness’ (a measure of its reproductive success) – tells us about how evolution happens. And studying the mechanisms of evolution is easier and faster when it’s done with micro-organisms.
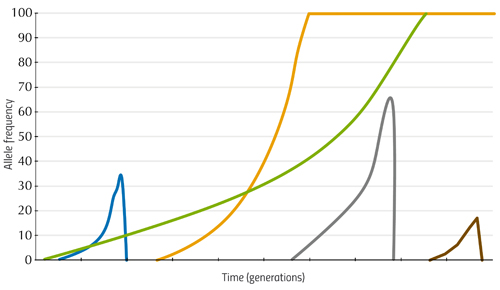
LTEE: a very long-term experiment
The experiment that aims to watch bacterial evolution in action is known as the E. coli long-term evolution experiment, or LTEE. In the experiment, which began on 24 February 1988, populations of E. coli are tracked over time to find out what changes take place in their genomes, and what impact these changes have on the organisms’ characteristics.
At the start of the experiment, Richard Lenski at Michigan State University, USA, set up 12 flasks with a bacterial medium containing the absolute minimum nutrients needed by bacteria to survive, then inoculated all 12 flasks with the same culture of E. coli, and left them incubating at 37 °C. The next day, he took out a small volume of the culture in each flask, diluted it with a fresh portion of the medium, and again left it to incubate overnight, discarding the previous day’s culture. This same process has been repeated every day since then – over 10 000 days so far.

Zachary Blount, Michigan State University
So how do the LTEE researchers find out whether the E. coli are evolving – and if so, how? Every 75 days (about 500 generations), a sample of the culture in each flask is frozen in a way that lets it be recovered, regrown on a Petri dish and analysed when needed. For each analysis, the researchers look at the DNA in the genomes of the different E. coli strains to map the genetic changes that occurred since the previous frozen generation in each flask. Crucially, they also compare how well strains from different flasks and time periods grow when competing with each other, because their growth rate is a good measure of evolutionary fitness: bacteria that grow and multiply faster are, in evolutionary terms, fitter.
From these analyses, Richard Lenski and his colleagues were able to establish the relationship between the mutations occurring in the E. coli genome and their effect on fitness – that is, they showed how the bacterium was evolving. Some of these findings are illustrated in figure 1.
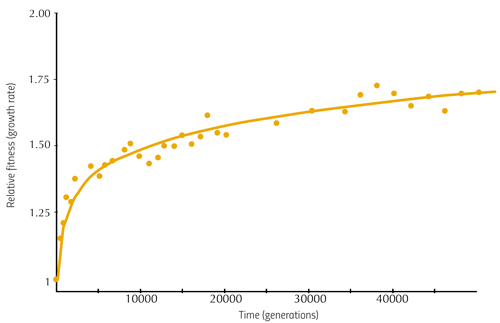
Here, the vertical axis shows the average growth rate of bacterial populations from six of the flasks, relative to the fitness of bacteria at the beginning of the experiment (i.e. how much better or worse the later cultures grow compared to the first), and the horizontal axis shows the generation number. We can see that in the earlier generations, the growth rate increases rapidly, indicating that the early mutations had a large effect. Later changes in DNA have a smaller influence, but the bacteria continue to adapt. There is no single ideal DNA sequence that provides perfect fitness in this environment, but rather a fitness-increasing path that the bacteria continue to climb.
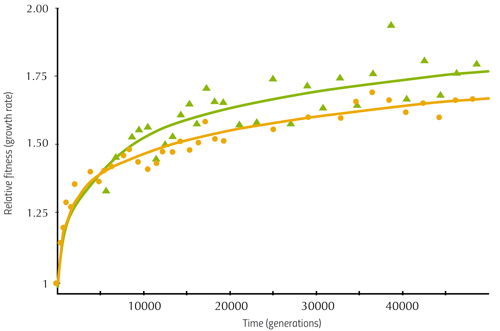
Another observation was that two unusual mutant strains emerged. One of them, which appeared independently in three populations between generations 2500 and 8500, was named the ‘hypermutator’ strain, because its DNA changed much faster than the DNA in other strains. The hypermutator strain achieved higher fitness sooner than other strains (figure 2), because more mutations – more genetic variants of the genome – meant that useful variants providing higher fitness were more likely to occur.
The second mutant that Lenski and his team discovered was even more impressive (Blount et al., 2012). Around generation 31 000, one strain started to grow much, much better than anything before it. Careful analysis of this mutant strain showed that it could use an alternative substance, citrate, as a key nutrient, in place of the small amount of glucose in the medium. The progeny of this mutant quickly took over the flask, and the mutation that enabled the bacteria to use citrate (see figure 3) became dominant in the population – a clear example of an advantageous mutation appearing and spreading throughout a population: positive natural selection in action.

Brian Baer and Neerja Hajela, Michigan State University
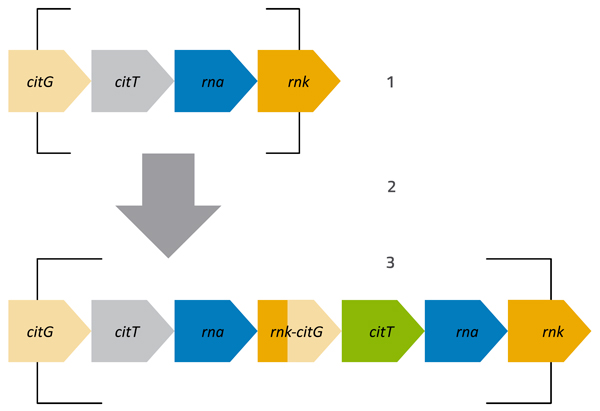
Stage 1: Original arrangement of the genome fragment, where citrate can only be used when no oxygen is present. The citT gene encodes a protein that transports citrate into the cell. When oxygen is present, this gene is inactive and the protein is not produced.
Stage 2: The genome fragment is duplicated.
Stage 3: After duplication, parts of the rnk and citG genes fuse. The activity of the rnk gene is not dependent on oxygen, and the activity of the rnk-citG fusion disrupts the control of the adjacent citT gene (green), activating it whether or not oxygen is present. The original copy of citT (grey) remains inactive in the presence of oxygen.
Learning from mutants
The citrate-eating mutant is a case in which it was possible to see evolution happening with the naked eye: a clear medium becoming cloudy with billions of cells literally swimming in a new source of food and growing explosively in number within hours. Although the initial mutation conferred only a small advantage over the previous generation of bacteria, it was enough to ensure the reproductive success of the mutants. Soon, the entire population was able to use citrate as a food source.
This is just one example of how the short generation time of micro-organisms, coupled with our new ability to sequence genomes very rapidly, allows us to see the effect of genetic variants playing out almost in real time. Studying these hallmarks of evolution in micro-organisms also enables scientists to comprehend evolutionary processes that take place in other organisms over much longer timescales – including the emergence of new species.
References
- Blount ZD et al. (2012) Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489: 513-518. doi: 10.1038/nature11514
- Wiser MJ, Ribeck N, Lenski RE (2013) Long-term dynamics of adaptation in asexual populations. Science 342: 1364-1367. doi: 10.1126/science.1243357
Resources
- Read an excellent discussion on how to define evolution.
- Richard Lenski’s blog entries about LTEE and related matters.
- Find out about evolution at the molecular level:
- Byrk J (2010) Natural selection at the molecular level. Science in School 14: 58-62.
- Byrk J (2010) Human evolution: testing the molecular basis. Science in School 17: 12-16.
- For Science in School articles on the anti-evolution idea, creationism, see:
- Leigh V (2008) Interview with Steve Jones: the threat of creationism. Science in School 9: 9-17.
- Pathmanathan S, Rau M (2008) Teaching in Sweden: tackling creationism, making waves. Science in School 10: 88-93.
Review
Teaching evolution is of vital importance in enabling students to become biologically literate citizens. Teachers are sometimes unable to convince their students of this important biological idea, so other interpretations (e.g. creationism) may find their way into the students’ thinking. This article can help teachers by providing new experimental results that support the theoretical concepts of evolution.
The article can be used in any classes relating to the study of evolution. Comprehension questions that students could be asked to answer include:
- How do organisms acquire diversity?
- Describe how genetic diversity is affected by natural selection.
- What does ‘fittest’ mean when used to describe organisms?
- What does this experiment tell us about the micro-evolution of organisms?
Panagiotis K. Stasinakis, biology teacher, 4th High School of Zografou, Greece





