Tara: an ocean odyssey Understand article
After four years travelling around the globe, the schooner Tara has returned with a world’s worth of scientific results.

City.
Image courtesy of
VHillaire/Tara Expeditions
It’s October 2011 and I’m on night duty aboard the schooner Tara as it glides across the Pacific Ocean’s dark and seemingly infinite waters. Tomorrow seems far off, but two things keep me awake: the smell of salt hanging in the air and specks of light scintillating in the wake of our boat. These ‘stars of the sea’ are in fact bioluminescent plankton – drifting micro-organisms so strange-looking that some of them inspired the design of creatures in the 1979 film Alien. Yet as tiny and bizarre as they may seem, plankton represent nine tenths of the living mass in the oceans and form the base of the global food web. Through photosynthesis, they generate half of the oxygen we breathe, draw carbon from the atmosphere into the deep sea, and play a crucial role in the global nitrogen cycle.

rosette that allowed them to
sample the oceanic plankton
at different depths.
Image courtesy of Tara
Expeditions
At dawn, the deck is abuzz with scientists who comb the upper ocean in search of plankton with thin nets, water pumps and a ‘rosette’, an instrument that traps water at different depths and measures its properties (mainly temperature, pressure and salinity). They catch all kinds of plankton, from tiny viruses 0.02 micrometres in diameter, to animals as ‘large’ as two millimetres across. This is roughly the ratio of the size of a golf ball to ten Olympic-sized swimming pools! Marine biologists funnel the specimens caught in the nets into test tubes, label them and freeze them to avoid chemical and enzyme degradation.
Down in the ‘dry lab’, a cabin filled with microscopes and computer screens, the imaging expert, Jérémie, places a drop of sampled water under a microscope. Suddenly, the boat is caught in waves that turn the entire lab into a swinging pendulum. I look for the edge of a table, anything, to keep my balance, while Jérémie, seemingly unaware of the complete havoc around us, sways in time with his microscope. He’s captivated by what he sees beneath the lens – this single droplet is teeming with improbable life forms….
An explosion of data
Over the course of Tara’s oceanic odyssey (2009–2013), more than two tonnes of frozen genetic material from plankton were shipped across the world to different laboratories for analysis. In the labs, researchers used chemicals to break open the specimens and extract their DNA molecules. They scanned the strands at an extremely high rate (a method known as shotgun sequencing) to generate a staggering list of 7.2 trillion pairs of nucleotides – the famous building blocks of DNA (adenine, thymine, guanine and cytosine) – and then used specific genes as ‘barcodes’ to identify different sorts of plankton, such as bacteria, Archaea and eukaryotes. Viruses, however, do not have a universal molecular identifier to be used as a barcode. Instead, researchers used protein clusters – groups of similar genetic sequences – to identify different viral populations.
Eric Karsenti, scientific director of the Tara Oceans project, explains the significance of this massive census. “The data we collected enable researchers to look in unprecedented detail at the populations, environments and dynamics of the oceans’ vital life support system.” He adds, “This is the first global description of the complete plankton ecosystem.”
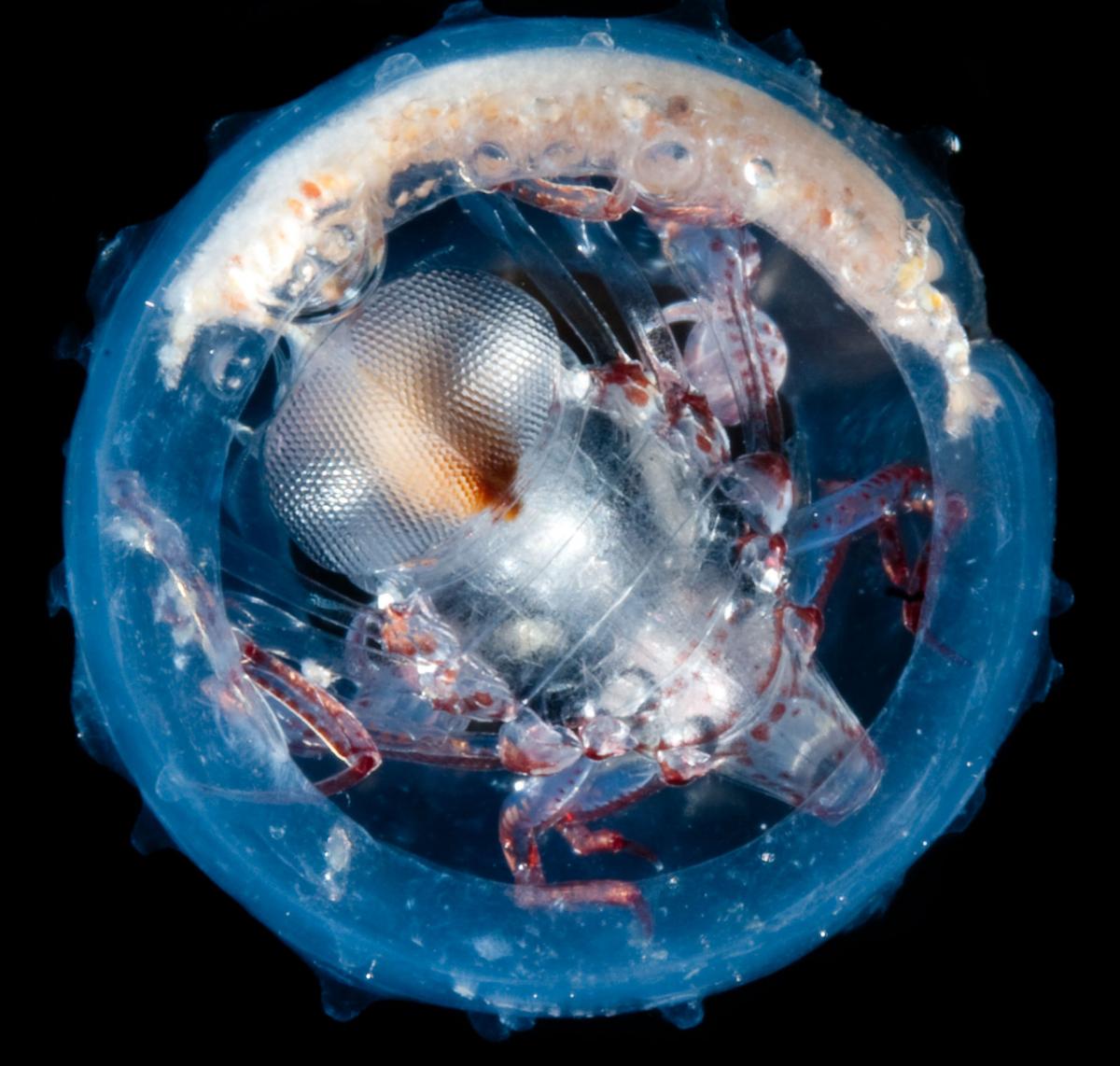
Image courtesy of M
Ormestad/Kahikai Tara Oceans
Experts from different fields analysed the sequenced data using advanced imaging, bioinformatics and the latest physical modelling technologies – techniques that are rarely used together. “This is the emergence of a new type of research in life sciences,” says Eric. “Five years ago, this was science fiction!” And together, the teams of researchers have begun to tackle questions that explorers of the past could not have even dreamed of addressing: What types of plankton populate our oceans? How do they interact with one another and their environment? How will they react to climate change and how will this affect us?
Back on dry land
The labs of the European Molecular Biology Laboratory in Heidelberg, Germany, might seem an unlikely place for ocean studies – they are a six-hour drive from the nearest coastline. But it’s here that Shinichi Sunagawa, a researcher in computational biology, helped create an ocean microbial gene catalogue of 40 million genes from microbial plankton, 80% of which are are new to science, indicating a huge biodiversity of unknown plankton in our oceans. Scientists found a strong correlation between the species that were found and the temperature of the habitat, identifying water temperature as the main environmental factor in shaping oceanic microbial communities. Further studies will determine how changes in water temperature could impact our oceans’ ecosystems and, consequently, our planet’s environment.

used to collect samples from
the surface.
Image courtesy of
V Hillaire/Tara Expeditions.
Most of the genes from Shinichi’s catalogue belong to eukaryotes – organisms (like us) whose DNA is coiled within a nucleus. This complex and stable cell structure was a milestone in evolution, enabling multicellular beings to form, and some eukaryotes have astounding properties as a result. Diatoms, for example, are single-celled organisms that synthesise a protective layer of glass at low temperatures, something we can only do using heat! Colomban de Vargas, a marine biologist who participated in both the expedition and the analyses, identified a total of 150 000 genetic types of eukaryotes – one hundred times more diversity than previously known. The key to this hyper-diversification lies in the species’ interactions.
An oceanic social network
On board Tara, scientists nicknamed the specimens they ‘met’ under the microscope: there was Hubert the protist and Dana the diatom. Later, Gipsi Lima-Mendez, a postdoc at the University of Leuven, Belgium, revealed the ‘social’ interactions between Hubert, Dana and their friends by helping create the oceanic interactome: a sort of planktonic Facebook that tells us which plankton are ‘friends” – always found together – and which are not. Then, she used computer-generated models to predict specific interactions between species, such as the symbiotic relationship between a flatworm and a photosynthetic microalgae: the microalgae lives inside the flatworm, safe from predators, and in exchange synthesises nutrients to feed its host. This predicted interaction was later observed using advanced microscopy of samples from the Tara expedition.

sampled by Tara in the north
Pacific.
Image courtesy of Eric
Roettinger / Kahikai / Tara
Oceans
Ocean interaction is far from ‘survival of the fittest’. According to Eric, “80% of interactions between organisms in the ocean are positive,” meaning that most organisms help one another to thrive. “This changes the way we look at evolution. Collaboration also makes life evolve and become more complex on Earth.”
The most abundant type of plankton is also the most elusive: viruses – so tiny that we could not see them with the microscopes on board the vessel. Ten million of them can squeeze into a single drop of seawater, and their impact is huge: they shape the populations they infect, drive evolution by transferring genes to different species, and “have global influences on the cycling of nutrients, organic matter and atmospheric gases”, says Jennifer Brum, a postdoc at the University of Arizona, USA, who took part in identifying more than 5000 viral populations, 99% of which were new. It’s like discovering a new underwater planet of alien life! The next step is to determine which viruses infect which organisms.
Collectively, these studies give us a benchmark against which to monitor the health of our oceans in the future. The 11.5 terabytes of data from the expedition – more data than Wikipedia – is stored at the European Nucleotide Archive, where it will remain in the public domain, available to current and future scientists. After all, scientists are still working with samples that Charles Darwin collected during his 1823 expedition on board the HMS Beagle. Who knows how long the Tara data will be answering questions that we haven’t even imagined yet.
More about EMBL
The European Molecular Biology Laboratory (EMBLw1) is one of the world’s top research institutions, dedicated to basic research in the life sciences. EMBL is international, innovative and interdisciplinary. Its employees from 60 nations have backgrounds including biology, physics, chemistry and computer science, and collaborate on research that covers the full spectrum of molecular biology. See: www.embl.org
EMBL is a member of EIROforumw2, the publisher of Science in School. Check out the list of all EMBL-related articles in Science in School.
Web References
- w1 – To learn more about EMBL.
- w2 – EIROforum is a collaboration between eight of Europe’s largest inter-governmental scientific research organisations, which combine their resources, facilities and expertise to support European science in reaching its full potential. As part of its education and outreach activities, EIROforum publishes Science in School.
Resources
- The non-profit organisation Tara Expeditions owns the schooner Tara and provides a lot of interesting and accessible resources on its scientific achievements on its website.
- More specifically, their pedagogical team has also set up projects and activities for schools.
- Read all the details and the most recent developments about the Tara Oceans project on the website of the European Molecular Biology Laboratory.
Institutions
Review
The Tara Oceans project brought together many fields of research that together produced the impressive Ocean Microbial Gene Catalogue, which will be used to monitor the health of our oceans.
The article offers the possibility to understand how scientists can characterise micro-organism populations and how environmental conditions shape ecological communities.
It can be used to study questions such as:
- What is the ecological role of plankton?
- Why are specimens frozen?
- What is DNA barcoding?
- What is the main environmental factor influencing ocean ecosystems?
- Why is the Oceanic Interactome compared to Facebook?
- What is the importance of viruses in ocean ecosystems?
Monica Menesini, Liceo Scientifico Vallisneri Lucca, Italy





