Shedding light on the gut microbiome Understand article
Learn how fluorescence microscopy can illuminate our gut microbiome and its role in cancer.
We live in a microbial world
A microbiome is a community of microorganisms present in any given environment. They inhabit nearly all habitats on the planet, from the upper atmosphere to the bottom of the ocean floor. They regulate carbon and nutrient cycling, animal (including human) and plant health, agriculture and the global food web.
Even our bodies are home to a vast microbiome consisting of trillions of bacteria, archaea, fungi and viruses. They help us digest our food, boost our immune system, produce vitamins[1] and even influence how we feel.[2] In fact, our microbiome is now considered an important organ we never knew we had.
The balancing act
Most of us dismiss microbes as germs that make us ill. In reality, most of our microbiome is friendly but some may become foes when disturbed. Let’s imagine our microbiome as a sea of friendly and not-so-friendly microbes. A sea is calm unless a hurricane strikes. A healthy microbiome exists in harmony unless threatened by an external force, such as a pathogen or antibiotics. When this happens, the balance tips, which can lead to an overgrowth of bad bacteria – a condition called as dysbiosis. Dysbiosis has been linked to diseases such as diabetes, obesity, irritable bowel syndrome and cancer.[3] What exactly happens when our microbiome goes out of whack?
Piecing together the puzzle: a look into colorectal cancer and its link to the gut microbiome
The gut microbiome is the largest microbiome in our body. Colorectal cancer (CRC), or cancer of the gut, has intrigued scientists for decades because it is linked to a severely dysbiotic gut microbiome. With the help of sequencing technologies, scientists have been able to compare the gut microbiome of healthy people with that of CRC patients. Their results unanimously show an overgrowth of certain not-so-friendly microbial groups in the guts of the cancer patients.[4,5] Some of these bacteria were even found within the tumours of CRC patients, which suggests a breach in the intestinal barrier,[6,7] the lining of mucus and cells on the walls of the large intestine that prevents microbes from attacking our bodies.
Whether dysbiosis is a cause or symptom of CRC remains an unsolved puzzle. Gene sequencing technologies can tell us what microbes are within any given sample, but not where they are. Understanding the location and interactions of CRC microbes with cells in the colon is crucial to know how microbes break the intestinal barrier, migrate into colonic tissue, and trigger cancerous signalling pathways that lead to the onset of CRC. Samples are broken down to extract nucleic acids prior to sequencing, which means that all information about location and tissue structure is lost. It is like acquiring all the pieces of a puzzle and trying to piece them back together in the dark.
Lighting up the gut microbiome
Fluorescence microscopy has the power to switch the lights on. By using a technique called fluorescence in-situ hybridisation (FISH), it is possible to light up different groups of microbes in different colours in a sample without destroying it. This is achieved by fluorescently labelling regions of bacterial ribosomal RNA that are unique to each group of bacteria.[8] The classic version of this technique uses one distinct fluorescent dye (fluorophore) per group.
Fluorescence microscopy
In fluorescence microscopy the sample is irradiated with light that matches the fluorophore excitation wavelength, which causes the fluorophore to emit light at a specific wavelength depending on its chemical structure. By filtering out the excitation light without blocking the emitted fluorescence, the fluorescence microscope selectively lights up the fluorescently labelled objects while the rest of the sample remains dark.
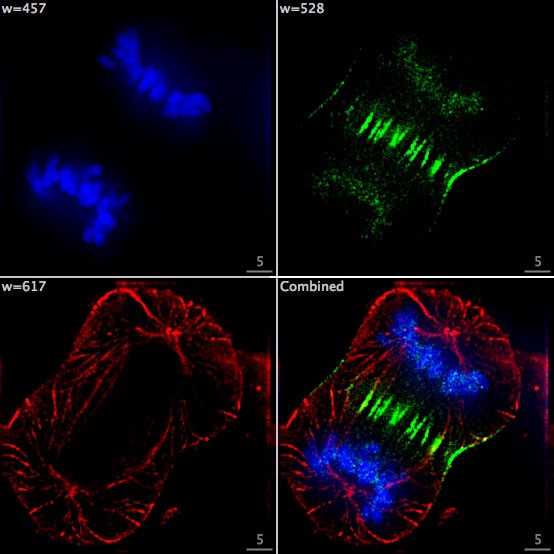
Image: F Lamiot/Wikimedia, CC BY-SA 4.0
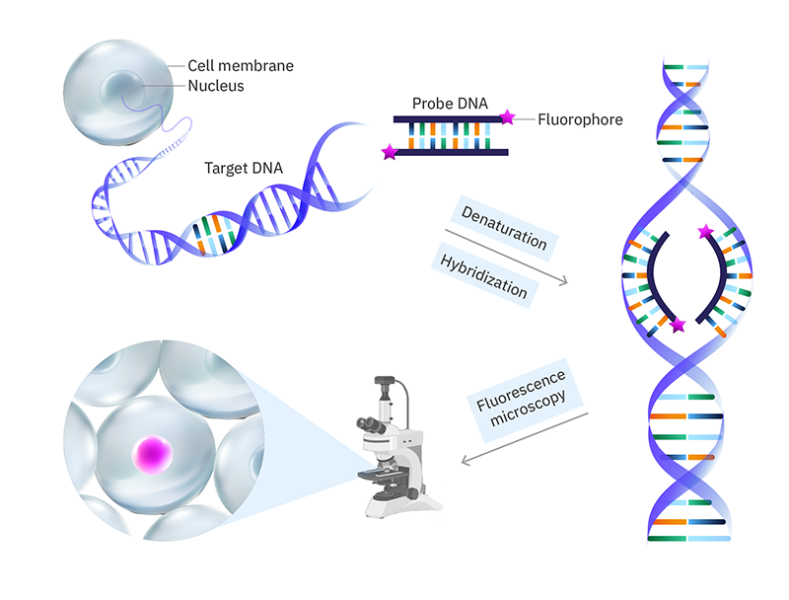
Fluorescence in-situ hybridisation (FISH)
FISH is a fluorescence labelling method in which fluorophores are attached to DNA sequences (called probes) that are complementary to a target nucleic acid (e.g., a portion of a gene). The cell walls (for bacteria) and lipid membranes in the sample are permeabilized so that the probes can get into the cell/nucleus. The DNA is then denatured (the two strands are separated) so that the probes can bind. The fluorescently labelled probes are then added and hybridized with their complementary sequences through base pairing. If the target nucleic acid is RNA, it is first reverse transcribed to DNA by using reverse transcriptase enzymes.
However, the visible-light spectrum has a limited number of easily distinguished colours and since the efficiency of a fluorescence microscope relies on its ability to filter the excitation light from the emitted light, visualizing more than four different fluorophore colours simultaneously becomes a challenge. This limited colour palette is not sufficient to label a complex CRC microbiome with hundreds of different microbial groups. So, scientists came up with a way to harness the power of advanced fluorescence microscopy. Each bacterial group is labelled with a unique DNA sequence called a barcode, like the barcode on our supermarket products, and then unique sequences of fluorophore colours are used to light up each group of microbes to scan the barcode and identify the bacterial group.[9] The number of possible permutations is huge so the number of bacterial groups we can identify is no longer limited by the number of distinguishable fluorophore colours. For example, with four fluorophore colours in a sequence of five, the possible permutations are 45=1024!
DNA-barcoded fluorescence microscopy
Instead of labelling the target nucleic acid with a single fluorophore, a specific DNA sequence called a barcode is used instead. Then to read the code, a set of fluorescently labelled detection probes is made for each barcode sequence. The barcode is decoded by using multiple cycles of hybridisation, with the detection probes removed before the next cycle. By choosing the colour of the detection probe for each barcode in each cycle and using a computer algorithm to combine the images at the end, you can assign each species a colour sequence to identify it in the sample.
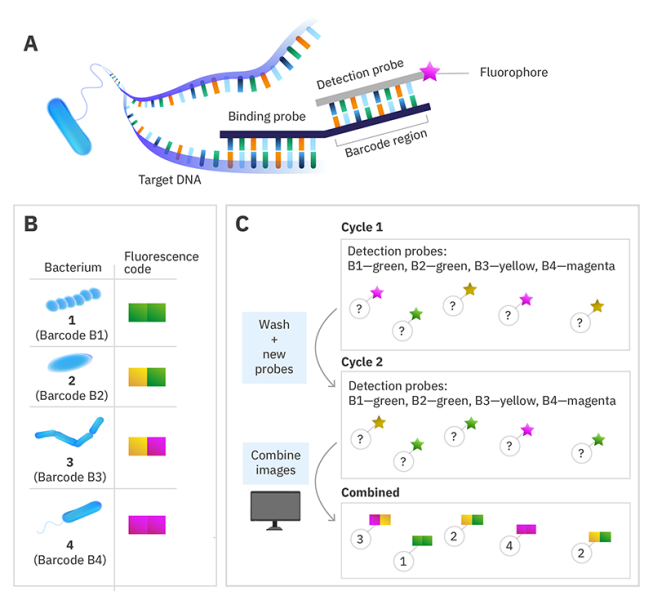
© Alexandra Krolik/EMBL, CC BY-NC 3.0 IGO
Using cutting-edge fluorescence microscopes, researchers at EMBL are now harnessing FISH to visualize hundreds or thousands of bacteria directly within a colon tumour. Bacterial groups relevant to CRC are assigned unique barcodes, fluorescently tagged with unique combinations of colours within the colon tumour tissue, and imaged on a fluorescence microscope. The images are decoded to reveal the identity and location of each bacterial group within the tumour.
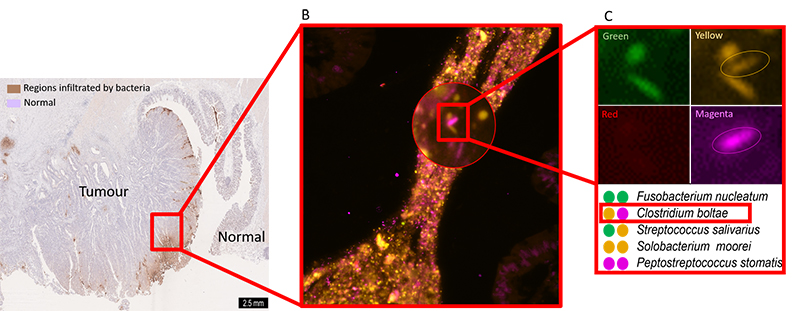
Image courtesy of Sebastian Degner
This barcoded FISH method can also be used to view cancerous colonic cells and immune cells gate-crashing the party! The puzzle starts to come together. But wait, it gets better: what if we could study the whole system of microbes infecting colonic cells in the lab? Scientists have successfully grown mini-colons on a plate and infected them with fluorescently labelled cancer-associated bacteria to observe the processes of infection in real time by using fluorescence microscopy. [10] This can potentially take the puzzle to near completion.
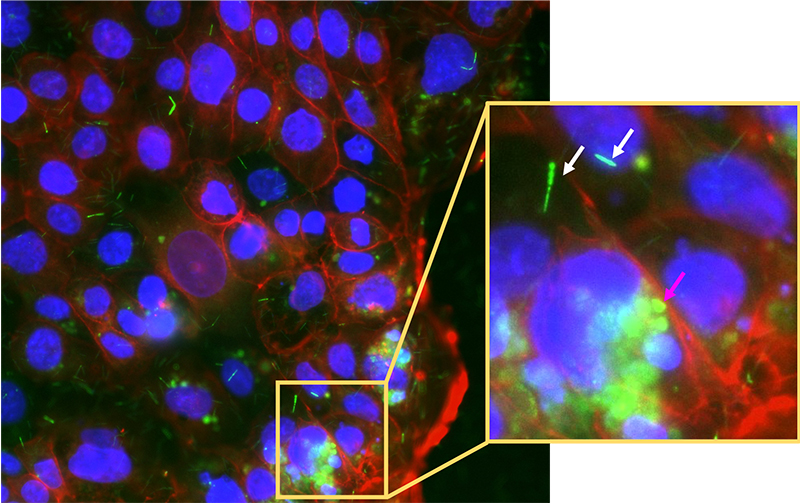
Image courtesy of the author
The exciting future of microbial fluorescence imaging
As amazing as it sounds, fluorescence microscopy does face some challenges. One major pitfall is the inability to distinguish true and false signals. False signals can come from fluorescent compounds inside a cell or artefacts introduced during sample processing. This issue can be tackled successfully with careful experimental design and efficient image-processing programs that can selectively remove background fluorescence. With its ability to illuminate the invisible world inside our guts, fluorescence microscopy has the power to visualize how microbes shape our bodies spatially, a dimension that is wholly absent in other approaches. An era of next-generation fluorescence microscopes with the ability to capture biological events in ultra-high definition within living organisms is approaching. It should soon be possible not only to visualize microbes colonizing your favourite model organ, but to zoom into each cell and witness cellular events simultaneously. It is like acquiring a cellular map with all necessary details to solve any biological puzzle. This enhanced understanding of how microbes contribute to cancer development could be used to develop new treatments or preventative measures.
References
[1] Kau AL (2011) Human nutrition, the gut microbiome and the immune system. Nature 474: 327–336. doi:10.1038/nature10213
[2] Pennisi E (2019) Gut bacteria linked to mental well-being and depression. Science 6427: 569. doi:10.1126/science.363.6427.569
[3] Carding S et al. Dysbiosis of the gut microbiota in disease. Microbial Ecology in Health and Diseases 26: 26191. doi:10.3402/mehd.v26.26191
[4] Wirbel J et al. (2019) Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nature Medicine 25: 679–689. doi:10.1038/s41591-019-0406-6
[5] Thomas AM et al. (2019) Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation Nature Medicine 25: 667–678. doi:10.1038/s41591-019-0405-7
[6] Bullman S, Kleger A (2017) Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358: 1443–1448. doi: 10.1126/science.aal5240
[7] Nejman D et al. (2020) The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368: 973–980. doi:10.1126/science.aay9189
[8] Pernthaler J et al. (2001) Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. In Paul J (ed) Marine Microbiology pp 207–226. Academic Press Ltd, London. ISBN: 9780125215305
[9] Valm AM, Welch JLM, Borisy GG (2012) CLASI-FISH: Principles of combinatorial labeling and spectral imaging. Systematic and Applied Microbiology 35: 496–502. doi:10.1016/j.syapm.2012.03.004
[10] Puschhof J et al. (2021) Intestinal organoid cocultures with microbes. Nature Protocols 16: 4633–4649. doi:10.1038/s41596-021-00589-z
Resources
- Watch this short video from the microbiology society about microbiomes.
- Watch a video about your own body’s microbiome.
- Read about Bacterial rRNA FISH
- Read about the DNA-barcoded FISH approach that was used in this research and some of its applications.
- Try out fluorescence microscopy in school using the microscopes in action kit: Arevalagam H, Gaikwad S (2022) Colours in the dark: fluorescence microscopy for the classroom. Science in School 58.
- Learn about the importance of animal use in research and some cutting-edge approaches to minimizing it: Schmerbeck S (2021) Organ-on-chip systems and the 3Rs. Science in School 54.
- Read about antibiotic resistance and drug development: Fernandez MD, Soler ML, Godinho T (2021) Microbiology: Discovering antibacterial agents. Science in School 55.
- Explore toxicology and the physiological effects of drugs using Daphnia as a model organism: Faria HM, Fonseca AP (2022) From drugs to climate change: hands-on experiments with Daphnia as a model organism. Science in School 59.
- Teach students how to collect relevant data regarding a gene from biological databases: Grazioli C, Viale G (2022) A chromosome walk. Science in School 57.
Institutions
Review
This is a very interesting and easy-to-read article that could be linked to important curriculum science topics such as microbiology, digestion, the cell cycle, and cancer.
In addition, it offers an exciting starting point for the Microscope in Action project presented in the recent Science School article Colours in the dark: fluorescence microscopy for the classroom, and could inspire more teachers to get involved. The Microscope in Action project, designed for teaching fluorescence microscopy in secondary-school classrooms, aims to develop content knowledge in biology and physics while also increasing the competencies of students in procedural knowledge, such as applying the scientific method and thinking in an interdisciplinary way. The current article provides a research example with real-world relevance to help put the hands-on activities in context.
Luis M. Aires, education researcher, School Group Antonio Gedeao, Portugal






