Bioinformatics: the new ‘cabinet of curiosities’ Understand article
The curation of ‘big data’ in molecular biology is changing the way scientists work.
In the 16th century, a cabinet of curiosities (or Wunderkammer) was a popular way to show off a private collection of extraordinary objects. Animal specimens, skeletons, minerals, unusual handmade objects and intriguing antiquities from the New World could all be revealed with a flourish, arousing in visitors a keen sense of curiosity in that new age of wonder.
Over time, cabinets of curiosities made way for more modern museums. Like the cabinets, museums catered for two profoundly human tendencies: curiosity, and the desire to collect and preserve knowledge.
Today, these same tendencies, coupled with new technology and a tsunami of genetic data, are driving a major change in the life sciences: the democratisation of access. As well as cataloguing the visible world of biological species, scientists can now sequence DNA from millions of species and enter the information into databases, along with other molecular biology data. The result is a new kind of menagerie: a constantly growing catalogue of biological information that can help scientists everywhere make sense of the living world.
But all this data needs curating, and the discipline of bioinformatics – which combines biology with computer science – has been developed to deal with this.

Spencer Phillips/EMBL-EBI
Opening up the cabinet
Research laboratories around the world produce a huge amount of data, which is then stored in specialised databases – such as those of the European Bioinformatics Institute (EMBL-EBI), located near Cambridge, UKw1. A key responsibility for EMBL-EBI is ensuring that the data it holds is publicly accessible, so the ‘collections’ are kept open to researchers everywhere. “It’s only in the past few years that this kind of openness has become workable, due to improved communication channels, but now it is expected by users”, says Andy Yates, a team leader at EMBL-EBI. “Data accessibility is crucial for anybody doing science. With a traditional cabinet of curiosities, the collector was the ultimate authority. We’re making the contents – and ourselves – open to reanalysis and review. It’s a necessary move if we want our resources to be truly useful”, he says.
Organising the data

the data centre houses vast
amounts of digital data,
using hundreds of servers.
EMBL-EBI
Traditional cabinets of curiosities organised items by type. The modern database organises biological data resources in a similar way – into categories. In the database, information and categories are interlinked, so the database is like a ‘smart’ or multidimensional cabinet of curiosities.
Indexing is as central to public data resources today as it was to earlier collections, to make data sets easy to find among the petabytes of data. Without indexing, there is no way of knowing what is in a database or how it got there. And descriptions of data sets – called metadata – are needed too: “Without metadata, exploring a database is like wandering through the basement of the Louvre blindfolded, hoping you’ll find the Mona Lisa”, says Yates.
To make these hard-earned data sets reusable by other scientists, data curators carefully check data submissions to ensure they meet the necessary requirements. These requirements are set out in widely accepted guidelines known by their abbreviation, FAIR: findable, accessible, interoperable and re-usable. Research data sets must also be put into context and linked to the scientific publication that describes them.
Visualising the data
Along with organisation, ways to visualise the data are also important: being able to ‘see’ connections within the data inspires people to keep exploring. “The first obvious difference between a cabinet of curiosities and a database is the content”, explains Jee-Hyub Kim, a former data miner at EMBL-EBI. “On the one hand, a collection of physical objects makes you feel something straight away. Just imagine what it must have felt like for someone who may have never even seen the ocean, to see and touch a starfish or coral. It’s difficult to create this sort of rapport with something as intangible as data. That’s why you need a good interface and visualisation tools – to allow the user to explore and interact with a data set or a digital object.”
One example of a data visualisation tool is the Protein Data Bank in Europe (PDBe)w2, a resource for collecting, organising and disseminating data on macromolecular structures, such as proteins. Apart from being a central repository for scientists studying proteins, PDBe allows users to see and interact with digital, three-dimensional models of proteins. These visualisations can be accessed from any internet-connected device throughout the world, including phones and tablets.
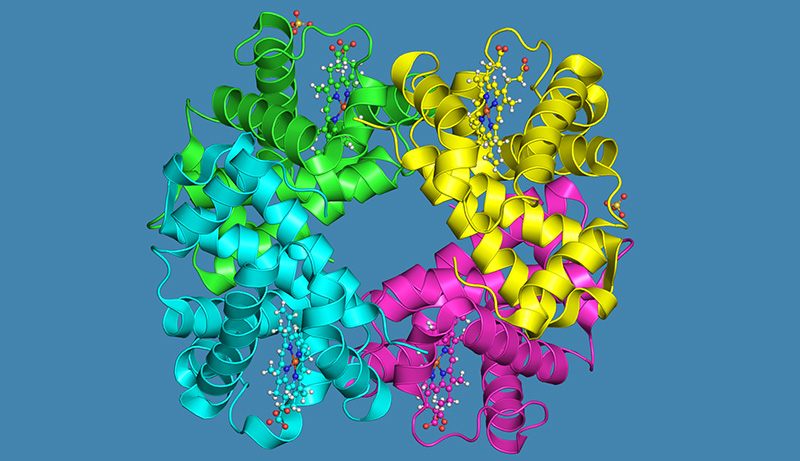
PDBe
New methods, new insights
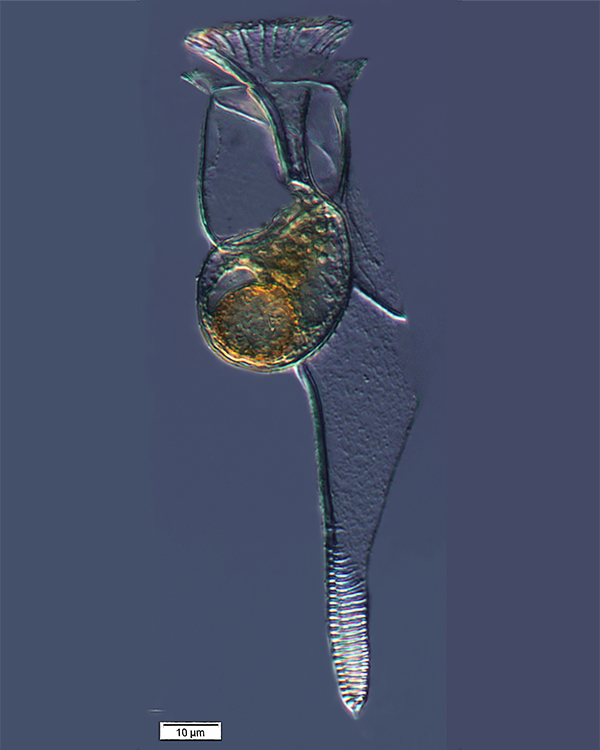
plankton Histioneis
elongata collected by the
Tara team in the South Pacific
Ocean
tintinnidguy/Flickr
So how is the availability of so much data changing the way we do science? According to Chuck Cook, scientific services manager at EMBL-EBI, scientists are going to become more dependent on big data – and those who don’t use big data will be left behind professionally. “As we become more specialised, running isolated experiments is becoming more difficult. To delve deeper into research, we will need to collaborate with people from lots of different backgrounds.”
“Biologists have to turn into programmers, to a certain extent”, agrees Yates. “That’s how the scientific questions are changing. The researcher will come up with a hypothesis and then prove or disprove it through data mining of large data resources. That requires some degree of programmatic knowledge.”
As scientists begin to analyse these data sets on a massive scale, they are revealing profound new insights. For example, the data from the Tara Oceans expeditions, in which a research ship has sailed more than 300 000 km worldwide since 2004, has led to the discovery of over 40 million new genes and is helping scientists to understand the invisible ecosystems that support the global food chain.
Scientists on the voyage systematically collected samples of plankton from all the world’s oceans, then shipped them back to land for DNA sequencing and analysis. “Sequencing the samples from Tara lets us ‘see’ some of the diversity of life in the oceans”, says Rob Finn, a team leader in EMBL-EBI’s metagenomics resource. “The first set of 40 million genes identified in Tara Oceans samples are mainly prokaryotes – bacterial species we haven’t seen before. But in the second wave of data, we have identified over 117 million eukaryote genes so far, and there is still a long way to go”, he says.
The nitty-gritty details
In light of this ever-growing influx of data, what are the big challenges facing biology in the coming years? “Before open data, a scientist worked on one protein, gene or experimental system, possibly for their entire career”, says senior scientist Janet Thornton, Director Emeritus of EMBL-EBI. “Seeing the bigger picture was practically impossible. Today, we can make genome-wide and species-wide observations”, she says. But Thornton thinks that this shift also poses the biggest challenge: truly important discoveries in biology still lie within the nitty-gritty details.
“We will still need to look closely at these details to understand many fundamental questions, such as why do organisms age?”, she says. “Initiatives like the Human Cell Atlasw3 are very good examples of all the missing details we still need to understand before we begin to explain how things work. The next step will be to translate this knowledge into everyday areas, such as medicine, agriculture and biodiversity.”
Much like the collectors who set up the first cabinets of curiosities, scientists are still meticulously cataloguing everything they learn about the form and function of life, and linking it all up to help them make further discoveries.
Acknowledgement
This article was adapted from the one originally posted on EMBL-EBI News, reproduced with kind permission.
Web References
- w1 – EMBL-EBI is the home of big data in biology. The institute hosts and shares data from life science experiments performed all over the world, and its scientists carry out basic research in computational biology. EMBL-EBI is one of the six sites of the European Molecular Biology Laboratory and is based just outside Cambridge, UK.
- w2 – PDBe is a database for three-dimensional structural data relating to large biological molecules, such as proteins and nucleic acids. The models are made freely available for scientists and students around the world.
- w3 – The Human Cell Atlas aims to map every single cell in the human body using single-cell sequencing technologies. This collaboration across the international scientific community brings together biologists, clinicians, geneticists, software engineers and others.
Resources
- Find out more about the Tara expeditions and ecological research on the Tara website.
- To read a Science in School article on the Tara expeditions, see:
- Peyrot R (2015) Tara: an ocean odyssey. Science in School 33: 6-11.
Institutions
Review
This article illustrates an idea that is fundamental for many disciplines, from natural science to economics: the huge amount of data and knowledge we now possess needs to be professionally organised so that it can be accessed by researchers all around the world.
In biology teaching, the article could be used to introduce the role of big data and bioinformatics in molecular biology, and to highlight how new computing technologies can help scientists to compare and visualise DNA and protein sequences. This could encourage students to explore for themselves the multiple possibilities that communication technologies are opening up in science.
The article could also be used to encourage awareness of the amazing biodiversity that has not yet been discovered in the oceans and other unexplored natural habitats.
Jesús López Alonso, biology teacher, IES La Gándara High School, Spain





