Neutrons and antifreeze: research into Arctic fish Understand article
Matthew Blakeley from ILL and his colleagues from ESRF and elsewhere have discovered how antifreeze in Arctic fish blood keeps them alive in sub-zero conditions. He and Eleanor Hayes explain.

(Myoxocephalus scorpius)
also has antifreeze proteins
Public domain image; image
source: Wikimedia Commons
When we think of the Arctic, many of us think of icebergs, polar bears and seals. Freezing temperatures, icy winds and desolate snowy wastes – a challenge to any animal that lives there.

forced animals to evolve
some remarkable adaptations
Image courtesy of ucumari;
image source: Flickr
We are familiar with many of the ways in which Arctic animals have adapted to their environment: the deep fur of polar bears, the thick blubber layer of seals, the migratory habits of many birds. These animals, however, are all endotherms – they maintain a constant body temperature, well above that of their icy surroundings.
For fish, the situation is very different – they are mostly exotherms, which means that their bodies are the same temperature as their surroundings. And their surroundings are very cold indeed: the Arctic Ocean frequently reaches temperatures as low as -1 °C, when it is only prevented from freezing by the high salt content of the water.
Fish blood, which is considerably less salty, would be expected to freeze at -0.5 °C. To avoid this, Arctic fish have evolved specialised antifreeze proteins (AFPs).

image source: Flickr
AFPs have a complex task. To prevent the fish from freezing to death, they must bind to ice-crystal nuclei forming in the fish’s body, preventing the nuclei from growing into ice crystals. How, though, do they distinguish ice from liquid water?
AFPs have been extensively studied using many techniques, including X-ray diffraction, revealing the existence of a specialised ice-binding surface.
X-ray diffraction relies on the degree to which X-rays are scattered (diffracted) by the electron cloud of each atom. Many molecules have distinctive diffraction patterns. However, hydrogen (and deuterium) atoms, which have only one electron, scatter X-rays very little (Figure 1a, below), so the X-ray diffraction signal for water (H2O) is much the same as for oxygen atoms (O). As a result, when scientists tried to examine how the ice-binding surface interacts with ice, they were unable to identify all the water molecules on the surface.
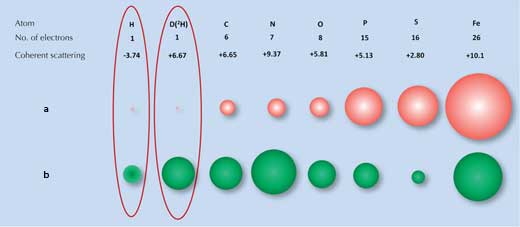
a) X-rays are scattered by electrons, with the strength of the scattering (size of the red balls)
being proportional to the number of electrons.
b) Neutrons are scattered by nuclei, with the strength of the scattering (size of the green balls)
depending on the specific nuclear forces
Image courtesy of Matthew Blakeley

antifreeze proteins of the
ocean pout, Zoarces
americanus
Image courtesy of Derek Keats;
image source: Flickr
This is where a second technique, neutron diffraction, is beneficial. Neutrons are scattered by atomic nuclei rather than electrons, and the strength of the scattering depends on the specific nuclear forces. These forces in turn depend not only on the elements but also on the elements’ isotopes, which differ in the number of neutrons in their nucleus (Figure 1b, above). Fortunately for us, hydrogen atoms can be easily detected by neutron scattering and the hydrogen isotope deuterium (heavy hydrogen, which has an extra neutron) can be detected even more reliably. Thus the signal for water is significantly different from that of oxygen.
We (Matthew Blakeley and his scientist colleagues) decided to investigate one type of AFP found in Arctic fish blood: type-III AFPs. To avoid the problems faced by other researchers, we used a combination of X-ray and neutron diffraction to examine the structure of the protein and its interactions with water molecules. For our experiments, we used the facilities at the Institute Laue-Langevin (ILL)w1 and the European Synchrotron Radiation Facility (ESRFw2; see box, below). The neutron research with such small crystal volumes was only possible due to the advances in sample preparation and instrumentation that have taken place at ILL over the past few years.
As is typical in investigations of protein structures, we inserted the synthetic gene for the type-III AFP into Escherichia coli, where it was over-expressed to allow us to collect sufficient amounts of the protein. The bacteria were supplied with heavy water (containing deuterium instead of normal hydrogen atoms), to ensure that the resulting protein was perdeuterated: the positions of all H atoms were occupied by deuterium. We then crystallised the protein so that we could determine its structure, checking that the perdeuterated form had the same structure as the normal form. (See Cornuéjols, 2009, and Blattmann & Sticher, 2009, to learn more about protein crystallography and even try it yourself.)
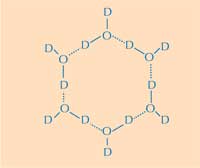
water ring. D = deuterium;
O = oxygen. Click on image
to enlarge
Image courtesy of Nicola Graf
Part of the answer to how type-III AFPs distinguish ice from liquid water lies in how the structure of the AFPs differs from that of typical proteins, which normally have hydrophobic amino acids in the core (away from water molecules in their surroundings) and hydrophilic amino acids on the surface. As has been shown by previous researchers, AFPs are unusual in having many hydrophobic amino acids on their surface, preventing the protein binding to liquid water via hydrogen bonds. These hydrophobic amino acids form part of the ice-binding surface, which binds to ice nuclei but not liquid water. How does this work?
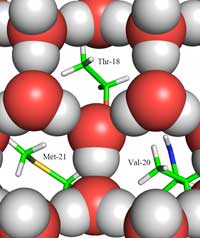
between ice (shown as red
and grey balls) and the ice-
binding site, showing the
methyl groups of the
hydrophobic residues
Thr-18, Val-20 and Met-21,
which face the holes in the
ice structure.
Click on image to enlarge
Copyright © 2011 John Wiley &
Sons, Ltd; image source:
Howard et al. (2011)
Herein lies the second part of the answer: how the structure of ice differs from that of liquid water. Using neutron diffraction, we were able to locate the positions of water molecules on the ice-binding surface. We identified a tetrahedral cluster of water molecules bound to the protein’s ice-binding surface. This tetrahedral cluster is found in liquid water (as in our experiment) but is also typical of ice; this gave us the starting point for modelling the rest of the ice crystal and deducing how it would bind to the AFP. The resulting model is composed of six-membered water rings in what is known as a boat configuration: six water molecules in a hexagonal arrangement, leaving a hole in the middle (see Figure 2, above). It is this hole that enables the type-III AFPs to distinguish the structure of ice-crystal nuclei from that of water: the hydrophobic regions of the ice-binding site fit into the holes, binding with the ice via Van der Waals forces (Figure 3, left). In contrast, liquid water has no hole into which hydrophobic regions such as methyl groups can fit. This prevents a large contact surface between liquid water and the ice-binding site, which would be necessary for a tight interaction.

the Arctic without antifreeze
proteins?
Image courtesy of dwleindecker
/ iStockphoto
Although other researchers have proposed that hydrophobic residues play an important role in how type-III AFPs recognise ice-crystal nuclei, this is the first experimental data that confirms it.
In itself, it is interesting to learn more about how Arctic fish survive in their environment, but this research also has potential industrial applications. Already, type-III AFPs are incorporated into some ice cream to minimise the build-up of large ice crystals and thus improve the consistency. In future, genes that code for AFPs may be incorporated into crops to allow them to be grown in sub-zero environments.
More about ILL and ESRF

The Institut Laue-Langevin (ILL)w1 is an international research centre at the leading edge of neutron science and technology. It operates one of the most intense neutron sources in the world, feeding beams of neutrons to a suite of 40 high-performance instruments that are constantly upgraded.
As a service institute, ILL makes its facilities and expertise available to visiting scientists. Every year, some 1200 researchers from more than 30 countries visit ILL in Grenoble, France. More than 800 experiments are performed every year, focused primarily on fundamental science in a variety of fields: condensed matter physics, chemistry, biology, nuclear physics and materials science.
The European Synchrotron Radiation Facility (ESRF)w2 is an international research centre, sharing a site with ILL in Grenoble, France. It produces high-brilliance X-ray beams, which serve thousands of scientists from all over the world, every year.
The complementarity of synchrotron X-ray beams and neutron beams can help us understand how complex systems work, such as the AFPs described in this article. To take full advantage of the synergy between synchrotron and neutron sciences, ESRF and ILL have been key actors in the creation of the Partnership for Structural Biology (PSB) and the Partnership for Soft-Condensed Matter; further partnerships are foreseen in the near future.
ILL and ESRF are members of EIROforumw3, the publisher of Science in School.
References
- Blattmann B, Sticher P (2009) Growing crystals from protein. Science in School 11: 30-36.
- Cornuéjols D (2009) Biological crystals: at the interface between physics, chemistry and biology. Science in School 11: 70-76.
- Howard EI, Blakeley MP et al. (2011) Neutron structure of type-III antifreeze protein allows the reconstruction of AFP–ice interface. Journal of Molecular Recognition 24: 724-732. doi: 10.1002/jmr.1130
Web References
- w1 – To learn more about ILL, see: www.ill.eu
- w2 – To find out more about ESRF and PSB, see www.esrf.eu and www.psb-grenoble.eu
- w3 – For more information about EIROforum, see: www.eiroforum.org
Institutions
Review
If you have ever wondered how fish can survive in icy oceans, you can find the answer in this highly engaging article, which leads the reader through a scientific investigation into the structure of proteins and their interactions.
The article could be used as background reading when studying biomolecules or as the basis of a discussion about the role of proteins in living organisms. Suitable comprehension questions include:
- Describe how Arctic fish are adapted to the icy Arctic waters.
- How do AFPs act as antifreeze in the fish’s blood?
- What are the limitations of X-ray diffraction when analysing AFPs?
- How does neutron diffraction differ from X-ray diffraction?
- Describe the steps involved in the successful examination of the AFP’s structure.
- How do AFPs differ from other proteins?
- How do AFPs distinguish ice from liquid water molecules?
Angela Charles, Malta





