Growing crystals from protein Teach article
Beat Blattmann and Patrick Sticher from the University of Zürich, Switzerland, explain the science behind protein crystallography and provide a protocol for growing your own crystals from protein – an essential method used by scientists to determine protein structures.

fragile objects, less than a
millimetre in diameter and
difficult to grow. Yet they are
essential for structural biology
studies by X-ray analysis
Image courtesy of Gaby Sennhauser
, University of Zürich
In 1959, Max Perutz and John Kendrew published an article on the three-dimensional structure of whale myoglobin, which is a small protein responsible for the transport of oxygen in whale cells. By investigating the protein’s structure, the two scientists wanted to understand the oxygen-carrying mechanism at the molecular level. They grew crystals from this protein and managed to determine its structure by analysing the X-ray diffraction pattern of the crystal.
A number of myoglobins from other species had been tested before with little success, until Perutz and Kendrew obtained a useable diffraction pattern with whale myoglobin crystals. This pioneering work was awarded the Nobel Prize in Chemistry in 1962w1. Fifty years later, however, it is still a challenge to obtain protein crystals for structural studies.
What are proteins?
Proteins are the largest group of non-aqueous components in living cells. Almost every biochemical reaction requires a specific protein, called an enzyme. Other types of proteins have mechanical and structural functions (e.g. collagen in connective tissue), or mediate cell signalling (e.g. hormone receptors), immune responses (e.g. antibodies) or the transport of small molecules (e.g. ion channels). The variety is immense: more than 20 000 different proteins are known to exist in humans alone.
Despite this variety, all proteins share an identical structural principle. They consist of 20 different building blocks, called amino acids, which are arranged in a linear chain connected by covalent bonds between adjacent amino acids (see figure below). The length of the protein chain varies from a few dozen to thousands of amino acids. In cells, each protein is assembled using the information encoded in its corresponding gene. The assembly is performed by a ribosome, which is a complex molecular machinery consisting of proteins and RNA.
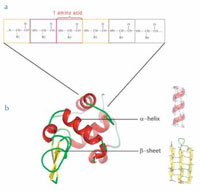
amino acids, which are
covalently linked to form a
linear chain
b. Proteins are folded to a
three-dimensional structure
that determines their
function. Small stretches of
the amino-acid chain form
typical folds. Two prominent
structural elements are
α-helices and β-sheets.
Click to enlarge image
Image courtesy of Marc
Leibundgut, ETH Zürich, and
www.pdb.org
Proteins are folded into distinct three-dimensional structures
Under natural conditions, the linear chains of amino acids spontaneously fold into distinct three-dimensional structures. Stretches of amino acids form typical secondary structural elements. The most prominent elements are α-helices and β-sheets (see figure below), which are typically stabilised by hydrogen bonds between individual amino-acid residues. The entire protein forms a tertiary structure consisting of a variety of such structure elements.
Structure is function: what does the three-dimensional structure of a protein tell us?
The function of a particular protein depends on its three-dimensional structure. Only when the protein is folded, the specific amino acids of the protein are close enough to enable the formation of an active site. These sites can catalyse biochemical reactions, as in the case of enzymes, or form a specific binding site, as in the case of antibodies. Investigating the structural details of a protein is of great importance to understand how fundamental processes of life function at a molecular level: this is the research area of structural biologists. One of the major challenges in structural biology today is the elucidation of the structure, function and interaction of huge macromolecular complexes and membrane proteinsw2. Due to their complexity, these proteins are experimentally extremely challenging, and every time the structure of a protein is determined, it is a major achievement. Nevertheless, since they are involved in fundamental biological processes, there is a great interest in better understanding their structure and function, and scientists keep trying to crystallise them.
Image courtesy of Beat Blattmann and Patrick Sticher
Proteins are too small for direct observation
Proteins are tiny structures, measuring only a few nanometres (1 nm = 1 millionth of a mm). Particles that size cannot be observed even with the strongest light microscope, which has a maximum resolution of 1 micrometre (1 m = 1 thousandth of a mm). Three major technologies are available to make protein structures ‘visible’:
- X-ray diffraction of protein crystals
- Nuclear magnetic resonance (NMR)
- Electron crystallography
As more than 90% of all protein structures deposited in the publicly accessible protein database of biological macromoleculesw3 have been determined by X-ray diffraction, we will concentrate on this method. To learn more about the history of crystallography and the journey of a protein from lab to lab, until its structure is solved, see the article by Dominique Cornuéjols in this issue.
Crystallising proteins is a tricky task, because it is difficult to determine the right conditions under which each new protein will crystallise – sometimes, it even seems impossible. So to ensure reproducible crystal quality (i.e. that equally good crystals can be grown again), scientists use controlled experimental set-ups to crystallise their proteins. The most frequently used method in protein crystallography is the vapour diffusion method (see image): in this method, a small amount of a crystallisation solution is added to the reservoir of the crystallisation chamber. A drop of protein solution and a drop of the crystallisation solution are pipetted onto the sitting drop post that is located in the centre of this chamber.
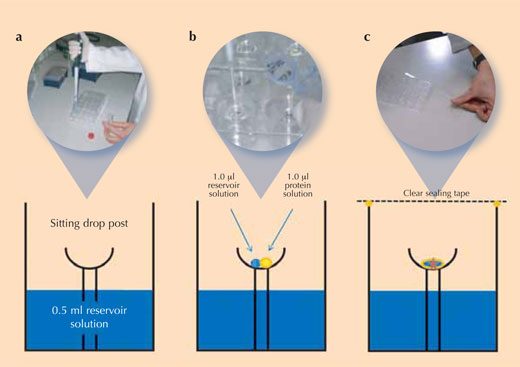
Image courtesy of Beat Blattmann and Patrick Sticher
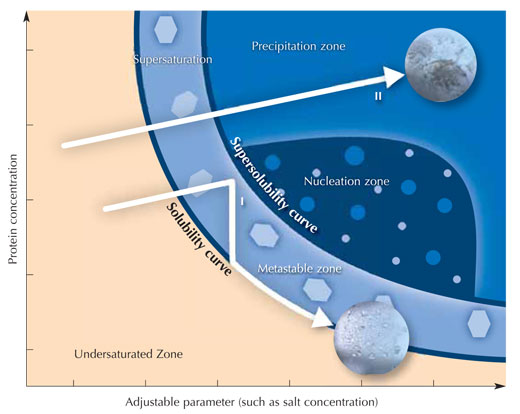
Immediately after adding all solutions, the chamber is sealed to avoid evaporation. Since the concentration of salt ions is higher in the crystallisation solution than in the mixture on the sitting drop post, solvent molecules will move from the protein drop to the reservoir by vapour diffusion in the gas phase. During this process, the solubility of the protein in the drop decreases. The protein solution in the drop eventually becomes supersaturated, which is a thermodynamically unstable state. This causes some of the protein in the drop either to form crystal nuclei that finally grow into larger protein crystals (see image), or to precipitate as amorphous protein which is useless for X-ray analysis. Crystallisation and precipitation are competing processes, so it is extremely important to find the optimal conditions favouring crystallisation.
Lysozyme crystals in the classroom
In this practical activity, students learn more about modern X-ray crystallography by determining the optimal crystallisation conditions for a protein. They investigate the formation of lysozyme crystals as a function of pH and salt concentration.
Lysozyme
Lysozyme is a protein belonging to a family of anti-bacterial enzymes which damage bacterial cell walls. In humans, it is abundant in a number of secretions, such as tears, saliva and mucus. Large amounts of lysozyme can also be found in chicken egg whites.
Equipment and materials
- One or two Cryschem™ crystallisation plates (Hampton Research) per class
- Crystal clear sealing tape (5 cm) (Hampton Research)
- 1 ml and 1 μl manual pipette
- A microscope to observe the crystals
- Storage space at 20 °C
Chemicals
- Lysozyme (SigmaAldrich Product #62971, BioChemika grade – lysozyme from a different source will probably also do, but this one has been thoroughly tested with the protocol, so it is recommended, to be on the safe side)
- Sodium chloride (NaCl) (table salt from the supermarket will do)
- Citric acid
- Sodium acetate
- Sodium phosphate, monobasic
- Sodium hydroxide solution
- Glacial acetic acid
- Deionised water (DI-water)
Stock solutions
The following aqueous stock solutions should be prepared in advance by the teacher:
- 50 mg/ml lysozyme stock solution in water
- 3 M sodium chloride
Dissolve 17.53 g NaCl in 100 ml DI-water. - 1 M sodium citrate, pH 3.5
Dissolve 19.24 g citric acid in 100 ml DI-water. Adjust the pH with sodium hydroxide solution to pH 3.5. - 1 M sodium acetate, pH 4.5
Dissolve 13.6 g sodium acetate in 100 ml DI-water. Adjust the pH with glacial acetic acid to pH 4.5 - 1 M sodium acetate, pH 5.5
Dissolve 13.6 g sodium acetate in 100 ml DI-water. Adjust the pH with glacial acetic acid to pH 5.5 - 1 M sodium phosphate, pH 6.5
Dissolve 15.6 g sodium phosphate in 100 ml DI-water. Adjust the pH with sodium hydroxide solution to pH 6.5.
Crystal growth experiment
- From the stock solutions, prepare the 24 reservoir solutions for the crystallisation experiments according to the table. The students can be split into small groups, each preparing some of the 24 different solutions. All groups can use the same stock solutions.
- Using the table for reference, pipette 0.5 ml of the corresponding reservoir solution into each of the 24 reservoir wells of a Cryschem™ plate (‘a’ in the figure above). The table summarises the conditions in each well and shows the position of the wells on the plate.
- Pipette 1 μl of the reservoir solution into the crystallisation cup on the sitting drop post in each well (‘b’ in the figure above)
- Add 1 μl of lysozyme stock solution to each 1 µl reservoir solution drop (‘b’ in the figure above).
- Immediately after adding the drops of protein solution, close the crystallisation vessel with crystal clear sealing tape to prevent evaporation from the vessel (‘c’ in the figure above)
- Store the plate at 20 °C. The crystals will start to grow immediately in some wells, and growth can be observed directly under the microscope at 1-2 hour intervals. The plates may be stored until the next lesson for final analysis. After about 1-2 weeks, crystals will have grown to their final size. A sealed plate will keep up to a year, sometimes even longer.
- Analyse the size, number and distribution of lysozyme crystals. The crystals may be too small to be observed with the naked eye, so a good magnifying glass or – even better – a microscope would be very useful.
- By comparing the results from the 24 reservoirs, determine the optimal conditions for crystallisati
| 1 | 2 | 3 | 4 | 5 | 6 | ||
|---|---|---|---|---|---|---|---|
| 2.0 ml of 3M NaCl stock solution (end conc. 0.6 M) 7.0 ml DI-water |
3.0 ml of 3M NaCl stock solution (end conc. 0.9 M) 6.0 ml DI-water |
4.0 ml of 3M NaCl stock solution (end conc. 1.2 M) 5.0 ml DI-water |
5.0 ml of 3M NaCl stock solution (end conc. 1.5 M) 4.0 ml DI-water |
6.0 ml of 3M NaCl stock solution (end conc. 1.8 M) 3.0 ml DI-water |
7.0 ml of 3M NaCl stock solution (end conc. 2.1 M) 2.0 ml DI-water |
||
| A | 1.0 ml sodium citrate (end conc. 0.1 M), pH 3.5 | A1 | A2 | A3 | A4 | A5 | A6 |
| B | B 1.0 ml sodium acetate (end conc. 0.1 M), pH 4.5 | B1 | B2 | B3 | B4 | B5 | B6 |
| C | 1.0 ml sodium acetate (end conc. 0.1 M), pH 5.5 | C1 | C2 | C3 | C4 | C5 | C6 |
| D | 1.0 ml sodium phosphate (end conc. 0.1 M), pH 6.5 | D1 | D2 | D3 | D4 | D5 | D6 |
Have your crystals measured by X-ray
When your class has successfully grown protein crystals, please contact Dr Patrick Sticher at sticher@bioc.uzh.ch. The Swiss NCCR (National Center of Competence in Research) Structural Biologyw2 has offered to produce an X-ray diffraction image for the first 10 school classes that successfully grow protein crystals using this protocol. X-ray measurements can be made either directly from school samples, or, if shipment is a problem, by reproducing the optimised crystallisation conditions found in your class and measuring those crystals. Together with the diffraction image, the scientists offer to send additional information on what they would do next with this information to obtain the actual structure, and a certificate if required.
Chat with scientists
Students can chat online with the scientists via Skypew4, after performing their own experiments. To make an appointment, email Patrick Sticher (sticher@bioc.uzh.ch) to chat with him using the Skype account ‘proteincrystallography’.
Download additional teaching material
A set of Powerpoint slides, images and further experiments are available onlinew5.
Suppliers
The following suppliersw6 provide the required materials and chemicals:
Hampton Research:
- Cryschem™ 24-1 SBS plate, Cat. No. HR1-002 (We recommend using this type of plate. One plate costs about US$3.)
- Crystal Clear Sealing Tape (5 cm), Cat. No. HR4-51
Gilson Inc:
- 1 ml and 1 μl manual pipettes
Sigma Aldrich:
- Lysozyme, Product #62971
- Sodium chloride, Product #71380
- Citric acid, Product #27488
- Sodium acetate, Product #71190
- Sodium phosphate, monobasic, Product #71502
References
- Cornuéjols D (2009) Biological crystals: at the interface between physics, chemistry and biology. Science in School 11: 70-76. www.scienceinschool.org/2009/issue11/crystallography
Web References
- w1 – Additional information about the 1962 Nobel laureates in chemistry and their pioneering work can be found on the website of the Nobel Prize Committee: http://nobelprize.org/nobel_prizes/chemistry/laureates/1962
- w2 – The Swiss National Center of Excellence in Research (NCCR) Structural Biology is a consortium of scientists dedicated to the elucidation of structure-function relationships of membrane proteins and supra-molecular complexes: www.structuralbiology.uzh.ch
- Selected research highlights can be found here: www.structuralbiology.uzh.ch/research004.asp
- w3 – New structures of biological macromolecules (proteins and nucleic acids) are deposited in the Protein DataBank (PDB). The website offers a number of interesting teaching resources: www.pdb.org
- Another valuable resource for protein information is: www.proteopedia.org
- w4 – To download and install Skype, see: www.skype.com
- w5 – Additional teaching resources are available here: www.structuralbiology.uzh.ch/teacher
- Login: crystallization
- Password: xraybeam2008
- w6 Hampton Research: www.Hamptonresearch.com
- Gilson Inc.: www.gilson.com
- Sigma-Aldrich: www.sigmaaldrich.com
Resources
Abad-Zapatero C (2002) Crystals and Life: A Personal Journey. La Jolla, CA, USA: International University Line. ISBN: 978-0972077408
Here are some recommended protocols for growing non-protein crystals with younger students:
- www.msm.cam.ac.uk/phase-trans/2002/crystal/a.html
- www.waynesthisandthat.com/crystals.
- htm http://chemistry.about.com/od/growingcrystals/Growing_Crystals.htm
Review
This article provides a good introduction to the study of protein crystals by X-ray diffraction. As such, it provides an interesting comprehension exercise for biology, chemistry and physics – showing good links between the three sciences. It can be used to discuss how to look at the very small, and why we need to study things at this level. The article also provides good background reading for teachers who are not aware of the use of diffraction as an analytical tool.
The practical looks like it will take a little time to set up and obtain results, but the offer of having the results analysed at a university gives it a different dimension to other practicals.
Mark Robertson, UK





