Locking the cradle Understand article
Winfried Weissenhorn’s group at the European Molecular Biology Laboratory in Grenoble, France, has uncovered a possible way to tackle a range of dangerous viruses –by trapping them inside their cocoons. Claire Ainsworth investigates.
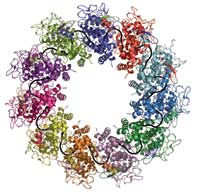
(in different colours) bind to one
RNA molecule (black) to form
a complex
Image courtesy of Winfried
Weissenhorn
It is perhaps one of the most feared diseases in the world. Once they appear, the classic symptoms – stumbling, drooling, fear of water, paralysis – almost always portend death. Only five people have ever survived the onset of rabies, and all but one suffered permanent neurological damage. Although vaccination and post-exposure preventive treatments have dramatically reduced the impact of the disease in Europe and North America, there is still no cure. Rabies remains a serious threat in many developing nations, killing up to 70 000 people every year.
Worryingly, rabies and similar viruses may now be re-emerging in developed countries, spread to people by contact with bats. So the discovery by Winfried Weissenhorn, Rob Ruigrok and their colleagues of an avenue to attack the virus is very timely. The teams, based at the European Molecular Biology Laboratory in Grenoble, France, and the neighbouring Unit for Virus Host Cell Interactions (UVHCI), have uncovered the structure of the protein that cocoons the virus’s genetic material and hides it from the body’s immune system until it has a chance to copy itself. It may be possible to use drugs to lock the genome inside this protective cradle, thus preventing viral replication.
But the work has implications beyond the realm of the single disease rabies. It turns out that many other viruses, including the Ebola, Borna disease and measles viruses, have similar cocoons for their genomes, meaning that this finding could also shed light on where such viruses came from and how they evolved. The rabies virus, like several other viruses, forms its genome out of a single strand of RNA, a molecule similar to the DNA that carries our genes. But unlike DNA, the information encoded by the virus’ RNA cannot be directly used to build the proteins needed to make the virus. Instead, it is a complementary copy, a kind of chemical photographic negative, of the sequence needed. So before the virus can make the proteins that constitute the shell protecting its genome, it first has to convert this ‘negative strand’ of RNA into ‘positive’ ones that can be translated into proteins. It also needs to make more copies of its genome to turn into more viruses.

Image courtesy of EMBL Photolab
For the virus, these processes are fraught with danger. Mammalian cells, including human cells, contain defence systems that attack and destroy foreign RNA. So the virus hides its vulnerable genome by packing it tightly inside a nucleocapsid, a shell made of a protein called nucleoprotein, to protect it until it can get inside the cell where it hijacks the host’s cellular machinery to replicate and produce viral proteins. As well as shielding the genome, nucleoprotein helps to control the balance between the production of proteins needed for viral replication and the process of replication itself, because both cannot happen simultaneously. In this way the nucleocapsid plays a key role in the virus’s life history.
Until now, however, the only clues scientists had about how nucleoproteins worked, came from fuzzy electron microscope images that showed how the nucleoprotein molecules polymerise on the genome to form nucleocapsids, but revealed little about the structure of the protein itself. To find out more, Rob Ruigrok of the UVHCI and Winfried Weissenhorn collaborated to make crystals of nucleoprotein and determine its structure using the high-intensity X-ray beams available at the European Synchrotron Radiation Facility, also in Grenoble. Crystals are symmetrical structures, and when exposed to X-ray beams they produce a highly ordered diffraction pattern from which scientists can deduce the precise shape of a molecule. By determining the structure of the nucleoprotein, the researchers would be able to start designing the drugs that could lock the viral genome within its protective shell.
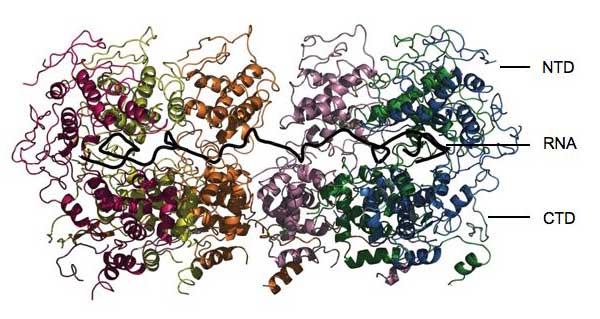
Image courtesy of Winfried Weissenhorn
The project began in Rob’s lab, where his team had been working on the nucleoproteins of a number of negative-strand RNA viruses since the mid-1990s. The rabies virus just happened to be the one that turned out to be the easiest to work with. Aurélie Albertini, a PhD student in the lab, had succeeded in getting insect cells in tissue culture to produce rabies nucleoprotein. The protein wrapped itself around the host cells’ RNA molecules, forming rings containing 9 – 13 protein molecules. Rob Ruigrok’s group had realised earlier that these behaved as miniature nucleocapsids, and electron microscopy studies provided coarse insights into their structure, which revealed that they closely resemble the rabies nucleoplasmid and hence could be used to study the structure of nucleoprotein arranged around RNA.
Aurélie started to work on producing nucleoprotein-RNA crystals, a project that was later joined by Amy Wernimont, a postdoc in Winfried’s lab. But the molecules were not playing ball, and both Aurélie and Amy struggled for a long time to get crystals that would allow Amy to determine the structure at a resolution of 4 Ångströms, which is sharp enough to distinguish how the protein folds, but not quite enough to show the sequence of amino acids, the individual building blocks of the protein. One problem was that the cells she was working with only produced tiny amounts of the nucleoprotein.
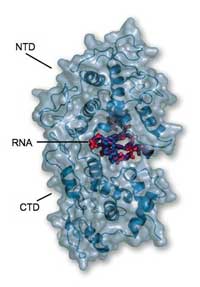
reveals that the RNA is completely
clamped at the interface of the
NTD (top) and the CTD (bottom)
and thus is not accessible to
degrading host enzymes or
the polymerase
Image courtesy of Winfried
Weissenhorn
Fortunately, Josan Márquez’s high-throughput crystallisation facility was at hand to help solve the problem, using only small protein samples. “The crystallisation robotics was absolutely wonderful,” recalls Winfried. “If we had needed large samples, we would not have been able to screen for the right crystallisation conditions.” The team eventually found the right conditions, the precise concentration of the protein and a complex mixture of chemical reagents needed for crystallisation, and managed to tweak them to gain 3.5 Ångström resolution – enough to construct a detailed model of the protein’s structure. Help also came from Raimond Ravelli, who assisted with data collection, fine-tuning the X-ray exposure time to get the best results and offering continuous advice at various stages of the investigation into the protein structure.
The results revealed that the nucleoprotein clamps itself completely around the RNA, locking it away like family jewels in a bank vault. “It’s not accessible for any other enzyme to attack it,” says Winfried. The protein is formed from two main working parts, or domains. One, called the CTD, cradles one side of the RNA and also sticks to the CTDs of other nucleoprotein molecules, helping to form a helical cocoon. The other domain, the NTD, sits on the other side of the RNA and does not make extensive contact with the other nucleoproteins. The overall structure is like a clamp squeezing around the RNA and keeping everything else out. “The nucleoprotein will prevent the RNA from being recognised by the innate immune system,” says Winfried. “But how does it become accessible for replication and translation?”
The answer lies in two stray-thread-like structures that protrude from either domain. These could act as hinges, swinging the NTD region up and bending the nucleoprotein clamp open, allowing viral enzymes access to small sections of the genome at a time. One particular protein, called phosphoprotein P, may be involved. It links RNA polymerase, the enzyme needed to copy the genome, to nucleoprotein, and may bind to one of the hinges to swing the NTD out of the way.
This hinge mechanism suggests a way to tackle viruses like the rabies virus with drugs that interfere with it. “If our concept that it opens up is correct, we could jam it shut,” says Winfried. “This would block viral replication.” Locked inside its protein cradle, the viral genome would be rendered powerless and eventually disposed of by the cell. The next step towards such drugs would be a systematic search for small chemicals with the ability to bind to the hinge region, thereby blocking the opening of the nucleocapsid.
The findings could also give some insight into how negative-strand RNA viruses evolved, says Winfried. Related virus species can have very different genome sequences, making it hard to draw any conclusions about their evolutionary history from sequence comparison alone. Structures, on the other hand, are a different story. The same physical structure can be built from a variety of gene and amino acid sequences. So even if genes evolve and change dramatically, the structures they encode can reveal deep evolutionary links between viruses.
Electron microscopy pictures of RNA molecules of other negative-strand RNA viruses, such as the measles virus, the Marburg virus and a crystal structure of a Borna virus, suggest that their nucleoproteins have a similar hinged clamp structure. This suggests these viruses use a tactic similar to the rabies virus’s for shielding their RNA, and so might also be targeted by drugs that jam their cocoons shut. It also suggests they share a common ancestor, says Winfried. “From the sequence analysis, you wouldn’t think they were related,” he adds. “I think there was probably some ancestral nucleocapsid, but then they diverged as the viruses evolved to infect different kinds of cells.”
Resources
- The work described in this article was published as:
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW (2006) Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313: 360-363.
Institutions
Review
This article illustrates the nature of scientific research as an ongoing, dynamic entity. As each molecular secret of the life cycle of even the simplest organisms is uncovered by use of sophisticated technologies, there are repercussions for advances in medical treatments and disease control.
Marie Walsh, Republic of Ireland





