A new tree of life Understand article
At the European Molecular Biology Laboratory in Heidelberg, Germany, Peer Bork’s research group has meticulously reconstructed a new tree of life – tracing the course of evolution. Russ Hodge explains.
In the margins of one of Charles Darwin’s notebooks is a small, twig-like drawing – unimpressive until you realise that it represents an enormous intellectual leap, a milestone in human history. It is the first modern sketch of a tree of life, representing the fact that distinct species had common ancestors. For a century, naturalists had collected facts about species, naming them and grouping them according to their similarities. Darwin suddenly understood that the similarities represented familial relationships.
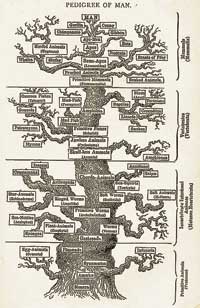
The Evolution of Man (1879)
Two decades later, another tree was meticulously composed by Ernst Haeckel, the great German naturalist and embryologist and a fanatical admirer of Darwin. Haeckel’s chart attempts to synthesize the plant and animal kingdoms into a single genealogical record of life on Earth. He got a lot of things right, but the tree goes back only so far. Once it reached one-celled organisms, he was stuck – scientists were only beginning to glimpse the amazing variety of such species alive on Earth; they certainly didn’t know enough to make a convincing phylogeny stretching back before the divergence of plants and animals.
Since then, scientists have filled in branches and twigs, climbed down the trunk, and pushed deeply into the roots, drawing on the written record of evolution that is preserved in DNA. Still, questions remain, particularly with regard to the early history of life on Earth. Peer Bork’s group at the European Molecular Biology Laboratory in Heidelberg, Germany, has now finished the highest-resolution tree of evolution that has yet been made. It may never be final – millions of species surely remain to be found, and those we know will continue to evolve. But it fills in many of the gaps, and will help scientists sort out fragmentary clues of the existence of new organisms. It also sheds light on the very early history of life on Earth.
Early in Earth’s history, there existed an organism that would give rise to all the species known today. In 1994, Christos Ouzounis and Nikos Kyrpides gave this shadowy creature a name: LUCA, for the last universal common ancestor. Studies of DNA sequences taken from plants, fungi, animals, bacteria, and another form of one-celled organism called Archaea proved that it must have existed. But until recently, scientists could say very little else about it.
“Two things have changed,” Peer says. “First is the immense amount of information we have from DNA sequencing – over 350 organisms have been completely sequenced, spread across the entire spectrum of life. This gives us a huge amount of data that can be compared to make a good tree and also to answer some questions about LUCA. Certain key genes can be found in all of them, and the chemical ‘spelling’ of these genes permits us to group them into families and historical relationships.”
It also allows researchers to reconstruct hypothetical ancestors. A fundamental principle of evolution, called the principle of common descent, states that if two organisms share features, it is almost always because they inherited the characteristics from a common ancestor. So by comparing existing species, scientists can obtain a picture of more ancient forms of life.
“Over the past few decades, scientists have realised there is an important exception to this rule,” Peer says. “Bacteria can swap genes with each other, and sometimes they can even steal a gene from a plant or an animal. Once that has happened, they pass the gene on to their descendents. Such genes have a completely different profile to genes inherited the normal way. It’s like finding a branch from a tree that grows crosswise and fuses into another branch.”
Peer says that attempts have been made to find such genes and eliminate them when building trees from DNA sequence data. But no one knew how often such events, called horizontal gene transfer (HGT), happened, or had developed a convincing method for finding them. “For a while, it was almost as if the amount of data was increasing the problem rather than solving it,” Peer says. “There were big debates, and the numbers of classifications were growing rather than reaching a consensus.” Part of the problem lay in the fact that the work could only be done by computer in a highly automated way, due to the incredible amount of genomic data that had to be sifted through.
Francesca Ciccarelli, a postdoc in Peer’s group, decided to tackle the problem of the tree anew and find a solution to the problem of the HGTs. She started by combing the complete genomes of 191 species for unique orthologues – genes in different species that had evolved from a common ancestral gene. The task was difficult because it couldn’t be completely automated. Francesca found 36 cases, five of which seemed to have been shuffled around through HGTs and were thus discarded.
Eliminating these from the analysis, the scientists could now build a complete tree by combining information from 31 genes. Peer was worried that some HGTs might have still have slipped in – a single mistake could spoil the quality of the tree. So the scientists put the computer to work doing some heavy lifting. The 31 genes were randomly divided into four groups. Trees were systematically drawn over and over again, for all of the genes in each group, with the exception of a single gene that was eliminated in each round. Then the results were compared. If the branches of the trees changed from pass to pass, an HGT was likely to be involved, and the gene was submitted to two more tests. In the end, the scientists found seven more candidates for HGTs, which they eliminated from their analysis.
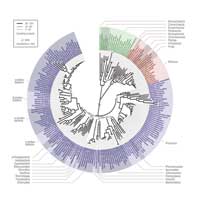
all three domains of life:
Archaea, Bacteria and
Eukaryota. There are so many
species that the tree has
to be drawn in a circle
The remaining information was combined into a super-tree which was compared once again to trees based on individual genes in three different ways. “Any one of these methods on its own might have left a tree with some mistakes,” Peer says, “but by combining them, we’re confident that we have an extremely accurate picture of the evolutionary history of these molecules and the species.”
The results clear up some old controversies, for example, a debate about the very early evolution of animals. Some trees in the past proposed that the vertebrates (which include humans) split off from another branch which would remain united for a while before splitting into separate branches leading to worms and insects. The new version groups things differently: vertebrate and insect ancestors split off from the worms together, and diverge from each other later.
The higher resolution of the tree is also important, Peer says, because of metagenomic studies which are underway to sequence all the genes found in environments such as farm soil or ocean water. His group has participated in several such projects. ”Most sequencing approaches start with a given organism and work through its whole genome systematically,” he says. “Metagenomics is sequencing a place – like a global positioning system coordinate. In many cases we recover fragmentary traces of thousands of genes, and have no idea what organism they come from. Often these molecules represent creatures that have never been seen before.” The breadth and detail of the new tree will allow scientists to make much better guesses about where such fragments fit in and what types of living beings they belong to.
Has the living world been fairly split up into major branches, limbs, and twigs, or have we overemphasized the prominence of our own lineage? A close look at the new tree shows that the latter seems to be the case. The eukaryotes, which include yeast, plants and animals such as ourselves, are so visibly different from one another that scientists have pushed them apart from each other on the tree. Genetically speaking, however, the species are often much more closely related than many single-celled forms of life.
“Smaller genomes evolve faster,” Peer says. “There isn’t a single organism that has been sequenced that is both evolving fast and has a large genome. It suggests that some of the simplest species around have ended up that way because they have pruned things down. Evolution isn’t always about acquiring complexity.”
The study also gives the scientists a closer look at LUCA. “One very big question has been what the earliest bacteria were like when they split off from the Archaea. Bacteria are grouped into two classes, called Gram-positive and Gram-negative, based on features of their membranes. The new tree reveals that Gram-positive bacteria evolved first. And if you look at their repertoire of genes, they seem to be suited to a very hot environment. The first Archaea were discovered in hot ocean vents, and most of the species alive today are thermophilic. It strongly suggests that LUCA was, too.”


